The IntFOLD Server Results
Please cite the following papers:McGuffin, L.J., Atkins, J., Salehe, B.R., Shuid, A.N. & Roche, D.B. (2015) IntFOLD: an integrated server for modelling protein structures and functions from amino acid sequences. Nucleic Acids Research, 43, W169-73. PubMed
Roche, D. B., Buenavista, M. T., Tetchner, S. J. & McGuffin, L. J. (2011) The IntFOLD server: an integrated web resource for protein fold recognition, 3D model quality assessment, intrinsic disorder prediction, domain prediction and ligand binding site prediction. Nucleic Acids Res., 39, W171-6. PubMed
Links to graphical output:
- Top 5 3D models
- Disorder prediction
- Domain boundary prediction
- Binding site prediction
- Full model quality assessment results
Download machine readable results in CASP format:
- TS (Tertiary Structure Prediction)
- DR (Disorder Prediction)
- DP (Domain Prediction)
- FN (Binding Site Prediction)
- QA (Model Quality Prediction)
Results will be available for 21 days after job completion (subject to server capacity)
| Top 5 3D models for T0515 | Help | ||||
| Model name (PDBsum links for templates used) | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | Model coloured by local quality (click images to view models, local errors and target coverage interactively) |
| nFOLD4_multi _HHsearch_TS1 2p3eA 2qghA |
CERT: 1.037E-4 |
0.6503 |
 |
 |
| nFOLD4_2qghA _HHsearch_TS2 |
CERT: 1.05E-4 |
0.6488 |
 |
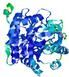 |
| nFOLD4_2qghA _COMA_TS6 |
CERT: 1.08E-4 |
0.6455 |
 |
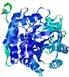 |
| nFOLD4_1twiA _COMA_TS3 |
CERT: 1.112E-4 |
0.6421 |
 |
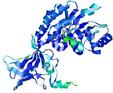 |
| nFOLD4_1twiA _HHsearch_TS5 |
CERT: 1.124E-4 |
0.6409 |
 |
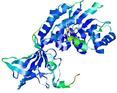 |
McGuffin, L. J. (2008) Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics, 24, 1789-1804. PubMed
| Disorder prediction for T0515 | Help | |||
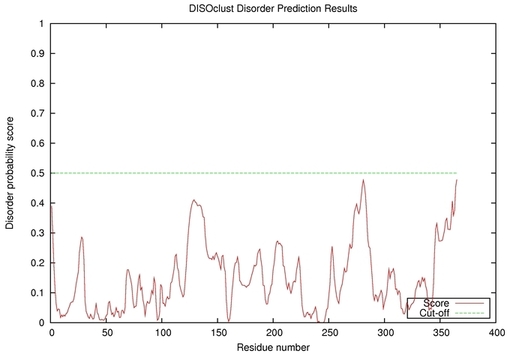
| |||
| Click image to download plot in PostScript format. |
| Domain boundary prediction for T0515 | Help | |||
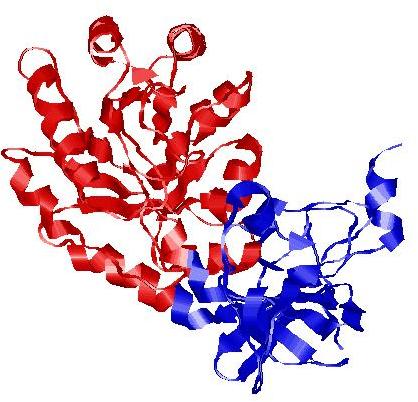
| |||
| Click image to download model or view prediction using Jmol. The model above is coloured according to the predicted domains. |
Roche, D. B., Tetchner, S. J. & McGuffin, L. J. (2011) FunFOLD: an improved automated method for the prediction of ligand binding residues using 3D models of proteins. BMC Bioinformatics, 12, 160. PubMed
| Ligand binding residues prediction for T0515 | Help | |||
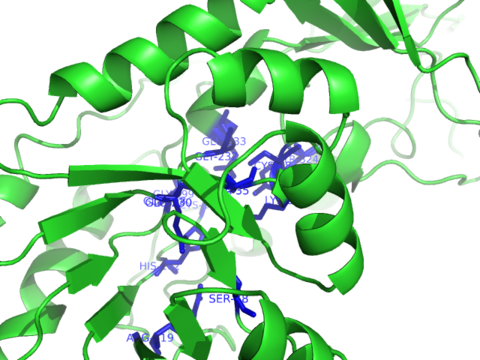
| |||
| Click image to download model or view prediction using Jmol. Predicted ligand binding residues are shown as blue sticks in the image above. |
McGuffin, L. J. & Roche, D. B. (2010) Rapid model quality assessment for protein structure predictions using the comparison of multiple models without structural alignments. Bioinformatics, 26, 182-188. PubMed
| Full model quality assessment results for T0515 | Help | ||||
| Model name (PDBsum links for templates used) | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | Model coloured by local quality (click images to view models, local errors and target coverage interactively) |
| nFOLD4_multi _HHsearch_TS1 2p3eA 2qghA |
CERT: 1.037E-4 |
0.6503 |
 |
 |
| nFOLD4_2qghA _HHsearch_TS2 |
CERT: 1.05E-4 |
0.6488 |
 |
 |
| nFOLD4_2qghA _COMA_TS6 |
CERT: 1.08E-4 |
0.6455 |
 |
 |
| nFOLD4_1twiA _COMA_TS3 |
CERT: 1.112E-4 |
0.6421 |
 |
 |
| nFOLD4_1twiA _HHsearch_TS5 |
CERT: 1.124E-4 |
0.6409 |
 |
 |
| nFOLD4_2p3eA _HHsearch_TS1 |
CERT: 1.172E-4 |
0.6360 |
 |
 |
| nFOLD4_2p3eA _COMA_TS2 |
CERT: 1.22E-4 |
0.6314 |
 |
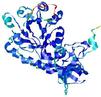 |
| nFOLD4_2j66A _HHsearch_TS3 |
CERT: 1.397E-4 |
0.6160 |
 |
 |
| nFOLD4_7odcA _HHsearch_TS10 |
CERT: 1.398E-4 |
0.6159 |
 |
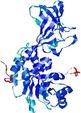 |
| nFOLD4_1tufa _sp3_TS2 |
CERT: 1.411E-4 |
0.6149 |
 |
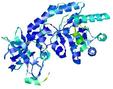 |
| nFOLD4_2o0tA _HHsearch_TS4 |
CERT: 1.458E-4 |
0.6112 |
 |
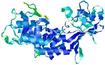 |
| nFOLD4_2oo0A _HHsearch_TS8 |
CERT: 1.486E-4 |
0.6091 |
 |
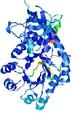 |
| nFOLD4_2j66A _COMA_TS5 |
CERT: 1.502E-4 |
0.6078 |
 |
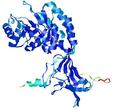 |
| nFOLD4_1knwA _HHsearch_TS6 |
CERT: 1.518E-4 |
0.6066 |
 |
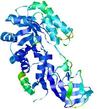 |
| nFOLD4_1knwA _COMA_TS9 |
CERT: 1.52E-4 |
0.6065 |
 |
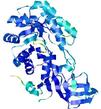 |
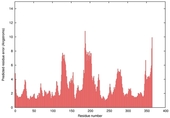
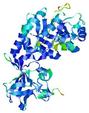
| nFOLD4_2yxxA _COMA_TS1 |
CERT: 1.523E-4 |
0.6063 |
 |
 |
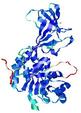
| nFOLD4_7odcA _COMA_TS10 |
CERT: 1.527E-4 |
0.6061 |
 |
 |
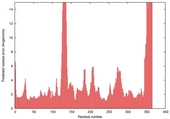
| nFOLD4_2pljA _HHsearch_TS9 |
CERT: 1.557E-4 |
0.6039 |
 |
 |
| nFOLD4_1f3tA _HHsearch_TS7 |
CERT: 1.558E-4 |
0.6038 |
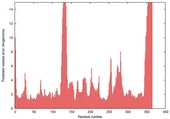 |
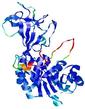 |
| nFOLD4_2yxxa _sp3_TS5 |
CERT: 1.665E-4 |
0.5964 |
 |
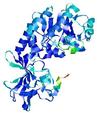 |
| nFOLD4_2todA _COMA_TS4 |
CERT: 1.68E-4 |
0.5955 |
 |
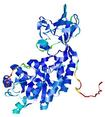 |
| nFOLD4_1hkva _sp3_TS3 |
CERT: 1.68E-4 |
0.5955 |
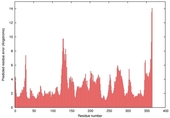 |
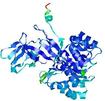 |
| nFOLD4_1hkva _spk2_TS3 |
CERT: 1.716E-4 |
0.5932 |
 |
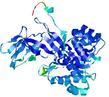 |
| nFOLD4_2nvaA _COMA_TS8 |
CERT: 1.813E-4 |
0.5872 |
 |
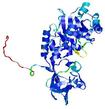 |
| nFOLD4_7odca _spk2_TS4 |
CERT: 1.85E-4 |
0.5850 |
 |
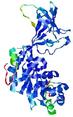 |
| nFOLD4_7odca _sp3_TS4 |
CERT: 1.871E-4 |
0.5838 |
 |
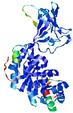 |
| nFOLD4_2plja _spk2_TS7 |
CERT: 1.892E-4 |
0.5826 |
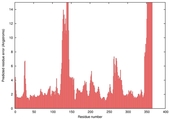 |
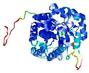 |
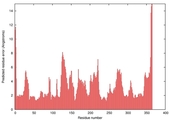
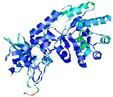
| nFOLD4_1tufa _spk2_TS2 |
CERT: 1.924E-4 |
0.5808 |
 |
 |
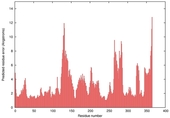
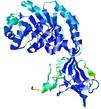
| nFOLD4_2j66a _sp3_TS6 |
CERT: 1.933E-4 |
0.5803 |
 |
 |
| nFOLD4_2plja _sp3_TS7 |
CERT: 1.934E-4 |
0.5803 |
 |
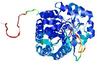 |
| nFOLD4_2j66a _spk2_TS6 |
CERT: 2.006E-4 |
0.5763 |
 |
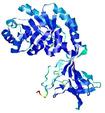 |
| nFOLD4_1knwa _sp3_TS1 |
CERT: 2.325E-4 |
0.5608 |
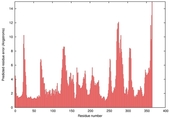 |
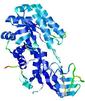 |
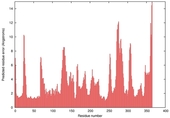
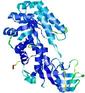
| nFOLD4_1knwa _spk2_TS1 |
CERT: 2.352E-4 |
0.5595 |
 |
 |
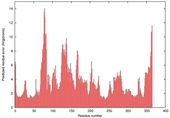
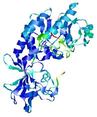
| nFOLD4_2yxxa _spk2_TS5 |
CERT: 2.998E-4 |
0.5347 |
 |
 |
| nFOLD4_3btnA _COMA_TS7 |
CERT: 3.262E-4 |
0.5264 |
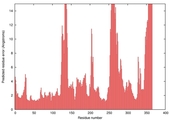 |
 |
| nFOLD4_7odca 2_sp3_TS9 |
HIGH: 1.696E-3 |
0.3820 |
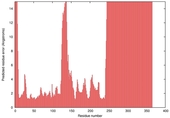 |
 |
| nFOLD4_7odca 2_spk2_TS9 |
HIGH: 2.004E-3 |
0.3693 |
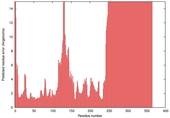 |
 |
| nFOLD4_3gwqa _sp3_TS8 |
HIGH: 2.216E-3 |
0.3618 |
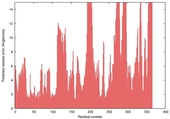 |
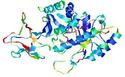 |
| nFOLD4_3gwqa _spk2_TS8 |
HIGH: 2.58E-3 |
0.3507 |
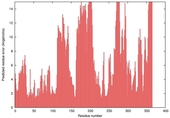 |
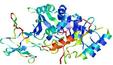 |
| nFOLD4_7odca 1_sp3_TS10 |
LOW: 9.248E-2 |
0.1494 |
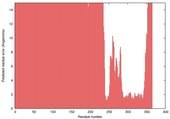 |
 |
| nFOLD4_7odca 1_spk2_TS10 |
POOR: 1.387E-1 |
0.1320 |
 |
 |