The nFOLD3 Protein Fold Recognition Server
NOTE: This server is no longer maintained, please use the latest version of IntFOLD to obtain predicted 3D models.
About the server
The nFOLD3 protein fold recognition server allows you to generate 3D models of a protein from its amino acid sequence. This new version of nFOLD uses the ModFOLD model quality assessment program to rank models built from several profile-profile alignment methods, which are run in-house - SP3, SPARKS, and HHsearch.
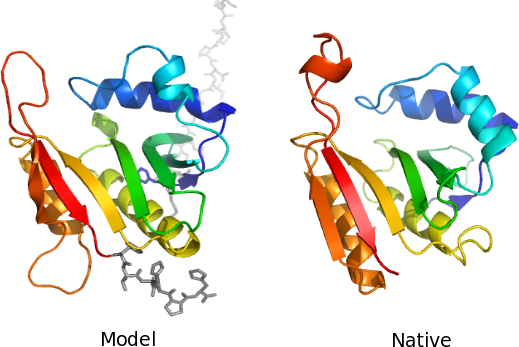
Figure 1 The nFOLD3 server has predicted the best model for CASP8 target T0417, compared with those predicted by all other servers (see Zhang's assessment). The nFOLD3 server model is on the left and the experimental structure is on the right. Note that the residues in the model represented as grey sticks are missing from the crystal structure. These residues were predicted to be disordered by the DISOclust server. Each image was rendered using Pymol and coloured according to the spectrum colouring option.
nFOLD3 Submission Form
News
- August 2011: The nFOLD3 server has been superseded by IntFOLD-TS method (nFOLD4) which is available as part of the IntFOLD server.
- March 2008: The new nFOLD3 server will be available in time for CASP8.
References
The original version of nFOLD was developed in 2004 at UCL in collaboration with Jaz Sodhi, Kevin Bryson and David Jones. A description of the method can be found in the following paper:
- Jones, D. T., Bryson K., Coleman, A., McGuffin, L. J., Sadowski, M. I., Sodhi, J. S. & Ward, J. J. (2005) Prediction of novel and analogous folds using fragment assembly and fold recognition. Proteins: Structure, Function and Bioinformatics, 61 Suppl 7, 143-51. PubMed
- McGuffin, L. J. (2008) The ModFOLD Server for the Quality Assessment of Protein Structural Models. Bioinformatics, 24, 586-587. PubMed
- McGuffin, L. J.(2007) Benchmarking consensus model quality assessment for protein fold recognition, BMC Bioinformatics, 8, 345. PubMed
- Söding J. (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics. 21, 951-96. PubMed
- Zhou H. & Zhou Y. (2005) SPARKS 2 and SP3 servers in CASP6. Proteins (Supplement CASP issue), Suppl 7 152-156.
Please also refer to the following papers which describe ModFOLD, SP3, SPARKS and HHsearch: