The IntFOLD Server Results
Please cite the following papers:McGuffin, L.J., Atkins, J., Salehe, B.R., Shuid, A.N. & Roche, D.B. (2015) IntFOLD: an integrated server for modelling protein structures and functions from amino acid sequences. Nucleic Acids Research, 43, W169-73. PubMed
Roche, D. B., Buenavista, M. T., Tetchner, S. J. & McGuffin, L. J. (2011) The IntFOLD server: an integrated web resource for protein fold recognition, 3D model quality assessment, intrinsic disorder prediction, domain prediction and ligand binding site prediction. Nucleic Acids Res., 39, W171-6. PubMed
Links to graphical output:
- Top 5 3D models
- Disorder prediction
- Domain boundary prediction
- Binding site prediction
- Full model quality assessment results
Download machine readable results in CASP format:
- TS (Tertiary Structure Prediction)
- DR (Disorder Prediction)
- DP (Domain Prediction)
- FN (Binding Site Prediction)
- QA (Model Quality Prediction)
Results will be available for 21 days after job completion (subject to server capacity)
| Top 5 3D models for T0635 | Help | ||||
| Model name (PDBsum links for templates used) | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | Model coloured by local quality (click images to view models, local errors and target coverage interactively) |
| nFOLD4_multi _HHsearch_TS1 3mn1A 3mmzA |
CERT: 1.333E-4 |
0.6212 |
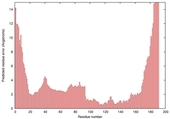 |
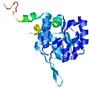 |
| nFOLD4_3mn1A _HHsearch_TS1 |
CERT: 1.363E-4 |
0.6187 |
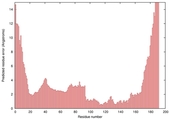 |
 |
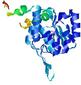
| nFOLD4_3mn1A _COMA_TS1 |
CERT: 1.381E-4 |
0.6172 |
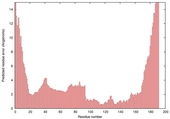 |
 |
| nFOLD4_3ij5A _HHsearch_TS3 |
CERT: 1.448E-4 |
0.6119 |
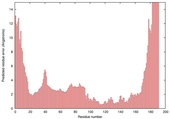 |
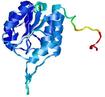 |
| nFOLD4_2r8eA _HHsearch_TS6 |
CERT: 1.504E-4 |
0.6077 |
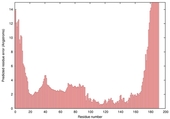 |
 |
McGuffin, L. J. (2008) Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics, 24, 1789-1804. PubMed
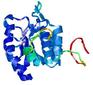
| Disorder prediction for T0635 | Help | |||
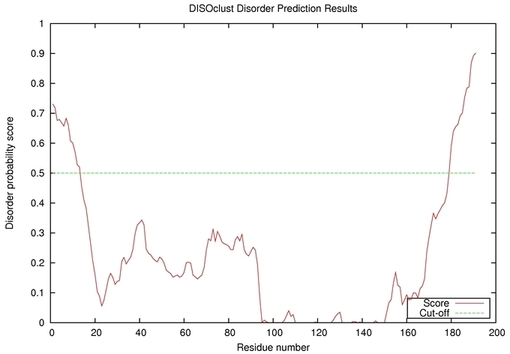
| |||
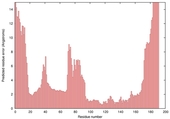
| Click image to download plot in PostScript format. |
| Domain boundary prediction for T0635 | Help | |||
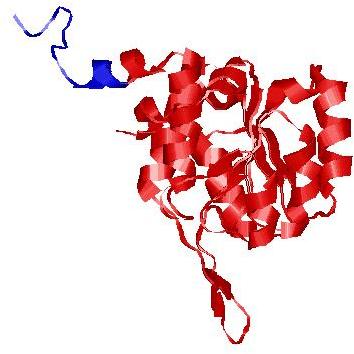
| |||
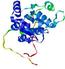
| Click image to download model or view prediction using Jmol. The model above is coloured according to the predicted domains. |
Roche, D. B., Tetchner, S. J. & McGuffin, L. J. (2011) FunFOLD: an improved automated method for the prediction of ligand binding residues using 3D models of proteins. BMC Bioinformatics, 12, 160. PubMed
| Ligand binding residues prediction for T0635 | Help | |||
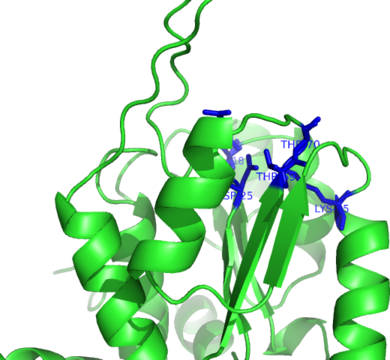
| |||
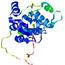
| Click image to download model or view prediction using Jmol. Predicted ligand binding residues are shown as blue sticks in the image above. |
McGuffin, L. J. & Roche, D. B. (2010) Rapid model quality assessment for protein structure predictions using the comparison of multiple models without structural alignments. Bioinformatics, 26, 182-188. PubMed
| Full model quality assessment results for T0635 | Help | ||||
| Model name (PDBsum links for templates used) | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | Model coloured by local quality (click images to view models, local errors and target coverage interactively) |
| nFOLD4_multi _HHsearch_TS1 3mn1A 3mmzA |
CERT: 1.333E-4 |
0.6212 |
 |
 |
| nFOLD4_3mn1A _HHsearch_TS1 |
CERT: 1.363E-4 |
0.6187 |
 |
 |
| nFOLD4_3mn1A _COMA_TS1 |
CERT: 1.381E-4 |
0.6172 |
 |
 |
| nFOLD4_3ij5A _HHsearch_TS3 |
CERT: 1.448E-4 |
0.6119 |
 |
 |
| nFOLD4_2r8eA _HHsearch_TS6 |
CERT: 1.504E-4 |
0.6077 |
 |
 |
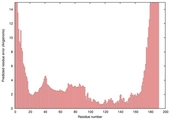
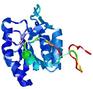
| nFOLD4_2r8eA _COMA_TS4 |
CERT: 1.505E-4 |
0.6077 |
 |
 |
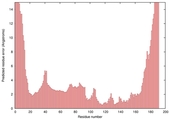
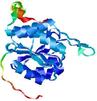
| nFOLD4_1k1ea _spk2_TS2 |
CERT: 1.612E-4 |
0.6000 |
 |
 |
| nFOLD4_1k1ea _sp3_TS1 |
CERT: 1.612E-4 |
0.6000 |
 |
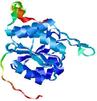 |
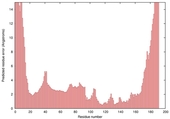
| nFOLD4_1k1eA _HHsearch_TS5 |
CERT: 1.634E-4 |
0.5985 |
 |
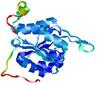 |
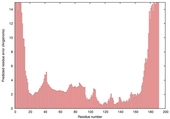
| nFOLD4_1k1eA _COMA_TS5 |
CERT: 1.667E-4 |
0.5964 |
 |
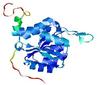 |
| nFOLD4_3mmzA _HHsearch_TS2 |
CERT: 1.72E-4 |
0.5929 |
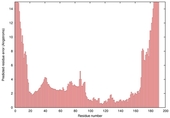 |
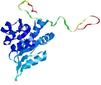 |
| nFOLD4_3mmzA _COMA_TS2 |
CERT: 1.72E-4 |
0.5929 |
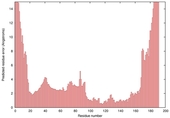 |
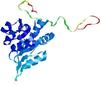 |
| nFOLD4_2p9jA _HHsearch_TS7 |
CERT: 1.736E-4 |
0.5919 |
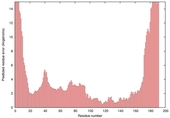 |
 |
| nFOLD4_3e81a _spk2_TS1 |
CERT: 1.779E-4 |
0.5892 |
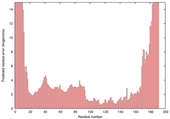 |
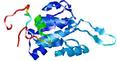 |
| nFOLD4_3e81a _sp3_TS2 |
CERT: 1.814E-4 |
0.5871 |
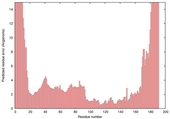 |
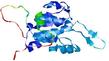 |
| nFOLD4_3e8mA _HHsearch_TS8 |
CERT: 1.836E-4 |
0.5859 |
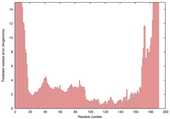 |
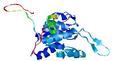 |
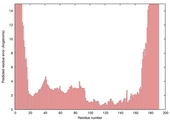
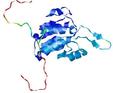
| nFOLD4_3e8mA _COMA_TS7 |
CERT: 1.851E-4 |
0.5849 |
 |
 |
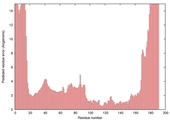
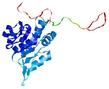
| nFOLD4_3mmza _sp3_TS4 |
CERT: 1.881E-4 |
0.5832 |
 |
 |
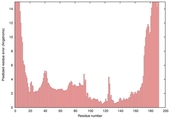
| nFOLD4_2p9ja _spk2_TS3 |
CERT: 1.911E-4 |
0.5815 |
 |
 |
| nFOLD4_2p9ja _sp3_TS3 |
CERT: 1.911E-4 |
0.5815 |
 |
 |
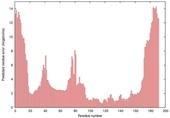
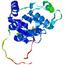
| nFOLD4_3ewiA _HHsearch_TS4 |
CERT: 1.956E-4 |
0.5790 |
 |
 |
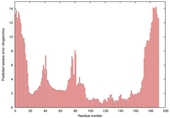
| nFOLD4_3ewiA _COMA_TS3 |
CERT: 1.956E-4 |
0.5790 |
 |
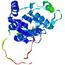 |
| nFOLD4_3mmza _spk2_TS4 |
CERT: 2.019E-4 |
0.5756 |
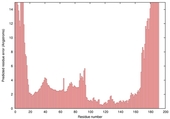 |
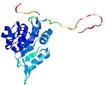 |
| nFOLD4_2p9jA _COMA_TS6 |
CERT: 2.16E-4 |
0.5685 |
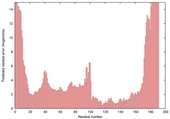 |
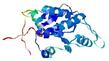 |
| nFOLD4_3ewia _sp3_TS5 |
CERT: 2.379E-4 |
0.5583 |
 |
 |
| nFOLD4_3ewia _spk2_TS5 |
CERT: 2.639E-4 |
0.5477 |
 |
 |
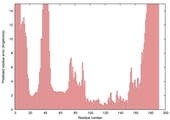
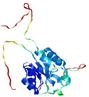
| nFOLD4_3fzqA _HHsearch_TS10 |
CERT: 4.961E-4 |
0.4862 |
 |
 |
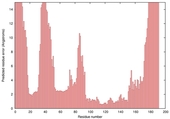
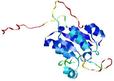
| nFOLD4_3dnpA _COMA_TS10 |
CERT: 5.052E-4 |
0.4844 |
 |
 |
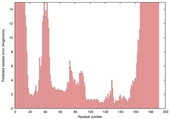
| nFOLD4_1l6rA _HHsearch_TS9 |
CERT: 6.137E-4 |
0.4667 |
 |
 |
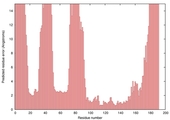
| nFOLD4_1wr8A _COMA_TS8 |
CERT: 7.135E-4 |
0.4533 |
 |
 |
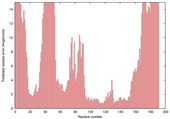
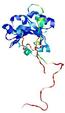
| nFOLD4_1l6rA _COMA_TS9 |
CERT: 9.112E-4 |
0.4322 |
 |
 |
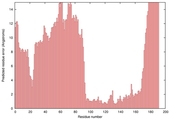
| nFOLD4_2b30a _spk2_TS7 |
HIGH: 1.442E-3 |
0.3947 |
 |
 |
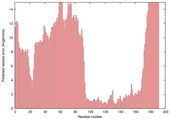
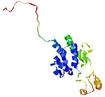
| nFOLD4_2b30a _sp3_TS7 |
HIGH: 1.442E-3 |
0.3947 |
 |
 |
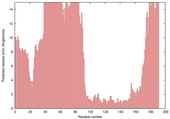
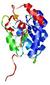
| nFOLD4_1wr8a _spk2_TS6 |
HIGH: 1.616E-3 |
0.3857 |
 |
 |
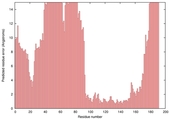
| nFOLD4_1wr8a _sp3_TS6 |
HIGH: 1.662E-3 |
0.3835 |
 |
 |
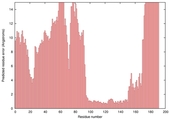
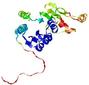
| nFOLD4_1rlma _spk2_TS9 |
HIGH: 1.755E-3 |
0.3794 |
 |
 |
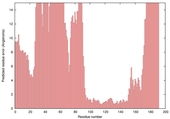
| nFOLD4_1rkqa _sp3_TS8 |
HIGH: 1.945E-3 |
0.3716 |
 |
 |
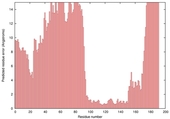
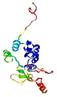
| nFOLD4_1rkqa _spk2_TS8 |
HIGH: 1.969E-3 |
0.3706 |
 |
 |
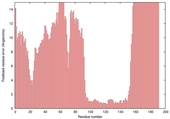
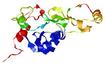
| nFOLD4_1rlma _sp3_TS10 |
HIGH: 2.829E-3 |
0.3440 |
 |
 |
| nFOLD4_3fzqa _spk2_TS10 |
HIGH: 2.869E-3 |
0.3431 |
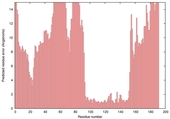 |
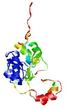 |
| nFOLD4_1nf2a _sp3_TS9 |
HIGH: 4.022E-3 |
0.3196 |
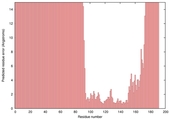 |
 |