The IntFOLD Server Results
Please cite the following papers:McGuffin, L.J., Atkins, J., Salehe, B.R., Shuid, A.N. & Roche, D.B. (2015) IntFOLD: an integrated server for modelling protein structures and functions from amino acid sequences. Nucleic Acids Research, 43, W169-73. PubMed
Roche, D. B., Buenavista, M. T., Tetchner, S. J. & McGuffin, L. J. (2011) The IntFOLD server: an integrated web resource for protein fold recognition, 3D model quality assessment, intrinsic disorder prediction, domain prediction and ligand binding site prediction. Nucleic Acids Res., 39, W171-6. PubMed
Links to graphical output:
- Top 5 3D models
- Disorder prediction
- Domain boundary prediction
- Binding site prediction
- Full model quality assessment results
Download machine readable results in CASP format:
- TS (Tertiary Structure Prediction)
- DR (Disorder Prediction)
- DP (Domain Prediction)
- FN (Binding Site Prediction)
- QA (Model Quality Prediction)
Results will be available for 21 days after job completion (subject to server capacity)
| Top 5 3D models for T0567 | Help | ||||
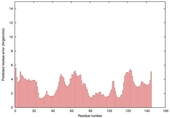
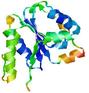
| Model name (PDBsum links for templates used) | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | Model coloured by local quality (click images to view models, local errors and target coverage interactively) |
| nFOLD4_multi _HHsearch_TS1 1ny5A 3dzdA |
CERT: 1.735E-4 |
0.5919 |
 |
 |
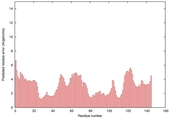
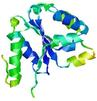
| nFOLD4_1ny5A _HHsearch_TS1 |
CERT: 1.813E-4 |
0.5872 |
 |
 |
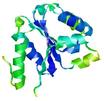
| nFOLD4_1ny5A _COMA_TS1 |
CERT: 1.856E-4 |
0.5846 |
 |
 |
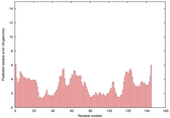
| nFOLD4_3dzdA _HHsearch_TS2 |
CERT: 1.939E-4 |
0.5799 |
 |
 |
| nFOLD4_3dzdA _COMA_TS2 |
CERT: 1.944E-4 |
0.5797 |
 |
 |
McGuffin, L. J. (2008) Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics, 24, 1789-1804. PubMed
| Disorder prediction for T0567 | Help | |||
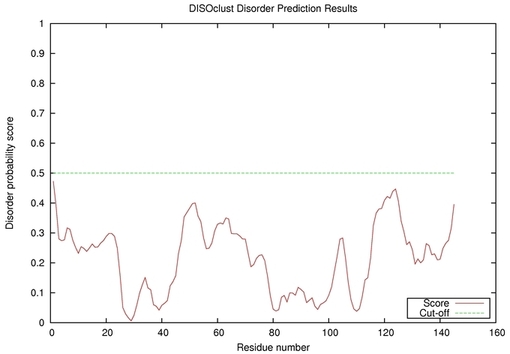
| |||
| Click image to download plot in PostScript format. |
| Domain boundary prediction for T0567 | Help | |||
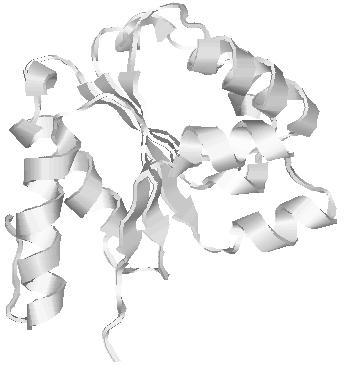
| |||
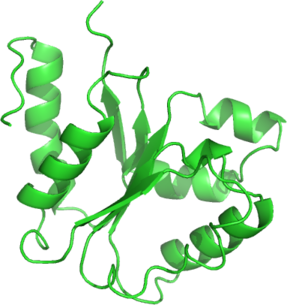
| Click image to download model or view prediction using Jmol. The model above is coloured according to the predicted domains. |
Roche, D. B., Tetchner, S. J. & McGuffin, L. J. (2011) FunFOLD: an improved automated method for the prediction of ligand binding residues using 3D models of proteins. BMC Bioinformatics, 12, 160. PubMed
| Ligand binding residues prediction for T0567 | Help | |||
| |||
| No binding residues predicted. |
McGuffin, L. J. & Roche, D. B. (2010) Rapid model quality assessment for protein structure predictions using the comparison of multiple models without structural alignments. Bioinformatics, 26, 182-188. PubMed
| Full model quality assessment results for T0567 | Help | ||||
| Model name (PDBsum links for templates used) | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | Model coloured by local quality (click images to view models, local errors and target coverage interactively) |
| nFOLD4_multi _HHsearch_TS1 1ny5A 3dzdA |
CERT: 1.735E-4 |
0.5919 |
 |
 |
| nFOLD4_1ny5A _HHsearch_TS1 |
CERT: 1.813E-4 |
0.5872 |
 |
 |
| nFOLD4_1ny5A _COMA_TS1 |
CERT: 1.856E-4 |
0.5846 |
 |
 |
| nFOLD4_3dzdA _HHsearch_TS2 |
CERT: 1.939E-4 |
0.5799 |
 |
 |
| nFOLD4_3dzdA _COMA_TS2 |
CERT: 1.944E-4 |
0.5797 |
 |
 |
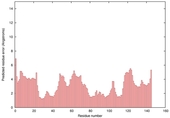
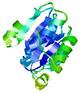
| nFOLD4_1ojlA _COMA_TS4 |
CERT: 1.983E-4 |
0.5775 |
 |
 |
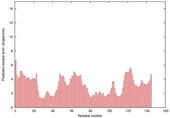
| nFOLD4_1ojlA _HHsearch_TS3 |
CERT: 2.017E-4 |
0.5757 |
 |
 |
| nFOLD4_2bjvA _HHsearch_TS4 |
CERT: 2.054E-4 |
0.5738 |
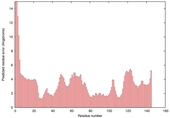 |
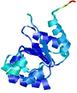 |
| nFOLD4_1ny5a _sp3_TS1 |
CERT: 2.066E-4 |
0.5732 |
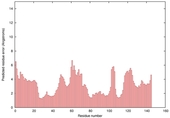 |
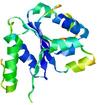 |
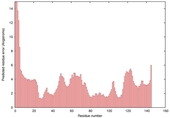
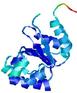
| nFOLD4_2bjvA _COMA_TS3 |
CERT: 2.091E-4 |
0.5719 |
 |
 |
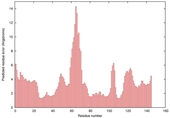
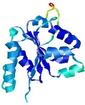
| nFOLD4_1ny5a _spk2_TS1 |
CERT: 2.349E-4 |
0.5597 |
 |
 |
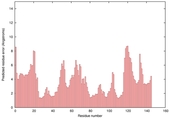
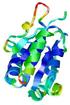
| nFOLD4_1iqpa 2_sp3_TS7 |
CERT: 3.407E-4 |
0.5221 |
 |
 |
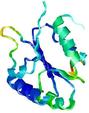
| nFOLD4_1sxje _sp3_TS5 |
CERT: 3.734E-4 |
0.5131 |
 |
 |
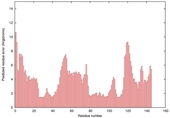
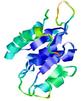
| nFOLD4_1a5t_ 2_spk2_TS3 |
CERT: 3.744E-4 |
0.5129 |
 |
 |
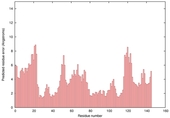
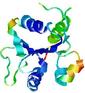
| nFOLD4_1qvrA _HHsearch_TS8 |
CERT: 3.762E-4 |
0.5124 |
 |
 |
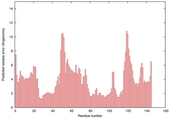
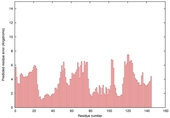
| nFOLD4_1sxje _spk2_TS10 |
CERT: 3.864E-4 |
0.5098 |
 |
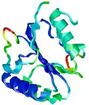 |
| nFOLD4_1sxjc _sp3_TS9 |
CERT: 3.968E-4 |
0.5073 |
 |
 |
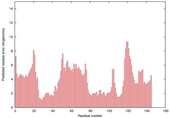
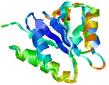
| nFOLD4_1fnna 2_sp3_TS8 |
CERT: 4.003E-4 |
0.5064 |
 |
 |
| nFOLD4_1a5t_ 2_sp3_TS3 |
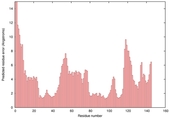
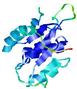
CERT: 4.003E-4 |
0.5064 |
 |
 |
| nFOLD4_1a5t_ _sp3_TS4 |
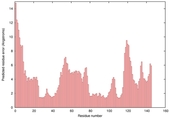
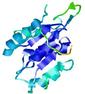
CERT: 4.022E-4 |
0.5059 |
 |
 |
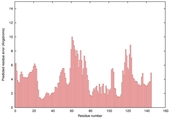
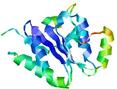
| nFOLD4_1fnna _spk2_TS9 |
CERT: 4.783E-4 |
0.4895 |
 |
 |
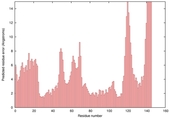
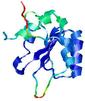
| nFOLD4_1g8pA _HHsearch_TS6 |
CERT: 5.759E-4 |
0.4724 |
 |
 |
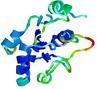
| nFOLD4_1njgA _COMA_TS10 |
CERT: 6.019E-4 |
0.4685 |
 |
 |
| nFOLD4_1g8pA _COMA_TS6 |
CERT: 6.035E-4 |
0.4682 |
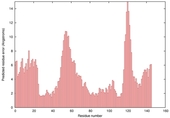 |
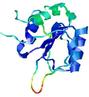 |
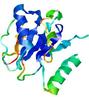
| nFOLD4_1jr3a 2_sp3_TS6 |
CERT: 6.212E-4 |
0.4656 |
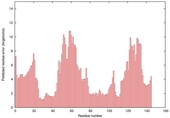 |
 |
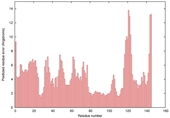
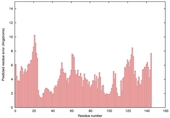
| nFOLD4_2r44A _COMA_TS7 |
CERT: 6.904E-4 |
0.4562 |
 |
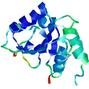 |
| nFOLD4_1jbka _sp3_TS10 |
CERT: 7.271E-4 |
0.4517 |
 |
 |
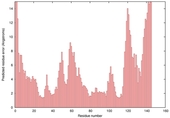
| nFOLD4_3co5a _sp3_TS2 |
CERT: 7.35E-4 |
0.4507 |
 |
 |
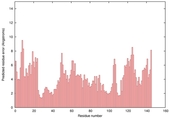
| nFOLD4_1jbka _spk2_TS5 |
CERT: 7.354E-4 |
0.4506 |
 |
 |
| nFOLD4_3co5a _spk2_TS2 |
CERT: 7.646E-4 |
0.4473 |
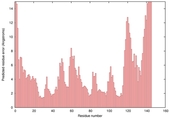 |
 |
| nFOLD4_1jbkA _COMA_TS9 |
CERT: 7.704E-4 |
0.4466 |
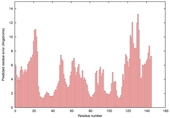 |
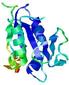 |
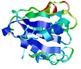
| nFOLD4_2p65A _COMA_TS8 |
CERT: 7.757E-4 |
0.4460 |
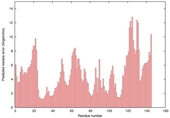 |
 |
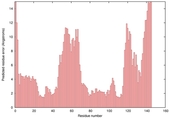
| nFOLD4_3co5A _HHsearch_TS5 |
CERT: 7.92E-4 |
0.4442 |
 |
 |
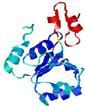
| nFOLD4_1in4a 2_spk2_TS6 |
CERT: 8.238E-4 |
0.4408 |
 |
 |
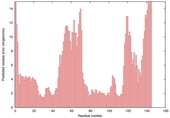
| nFOLD4_3co5A _COMA_TS5 |
CERT: 8.76E-4 |
0.4355 |
 |
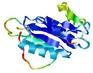 |
| nFOLD4_1fnna 2_spk2_TS4 |
CERT: 9.703E-4 |
0.4269 |
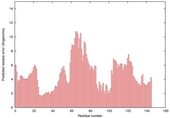 |
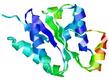 |
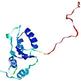
| nFOLD4_1d2nA _HHsearch_TS10 |
HIGH: 1.037E-3 |
0.4214 |
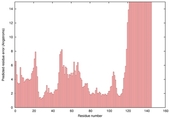 |
 |
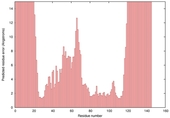
| nFOLD4_3f8tA _HHsearch_TS9 |
HIGH: 3.619E-3 |
0.3268 |
 |
 |
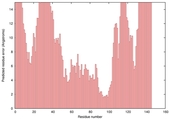
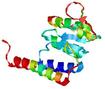
| nFOLD4_1lc0a 1_spk2_TS7 |
MEDIUM: 1.135E-2 |
0.2546 |
 |
 |
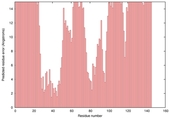
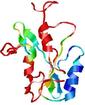
| nFOLD4_2eo6a _spk2_TS8 |
POOR: 1.705E-1 |
0.1235 |
 |
 |
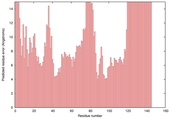
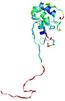
| nFOLD4_3cwoX _HHsearch_TS7 |
POOR: 2.278E-1 |
0.1116 |
 |
 |