IntFOLD: Integrated Protein Structure and Function Prediction
About the server
The IntFOLD server provides a unified interface that integrates:
- 3D Modelling: Comprehensive tertiary structure prediction and high-resolution model generation.
- Quality Assessment and Error Correction: Robust estimation of model accuracy (built-in self-estimates) coupled with automated structural refinement.
- Structural Analysis: Simultaneous prediction of intrinsically disordered regions and structural domain architecture.
- Functional Site Mapping: Precise identification of protein-ligand binding residues and potential interaction sites.
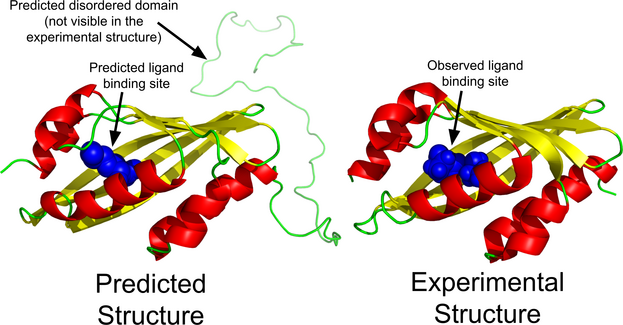
This website is free and open to all users and there is no login requirement.
Privacy and cookies
IntFOLD7 submission form
Help page
Sample output
Fair usage policy
You are only permitted to have 1 job running at a time for each IP address, so please wait until your previous job completes before submitting further data. If you already have a job running then you will be notified and your uploaded data will be deleted. Once your job has completed your IP address will be unlocked and you will be able to submit new data.
Latest IntFOLD server references
- McGuffin, L. J., Edmunds N. S., Genc, A. G., Alharbi, S. M. A., Salehe, B. R. and Adiyaman, R. (2023) Prediction of protein structures, functions and interactions using the IntFOLD7, MultiFOLD and ModFOLDdock servers. Nucleic Acids Research. gkad297. DOI PubMed
- McGuffin, L.J., Adiyaman, R., Maghrabi, A.H.A., Shuid, A.N., Brackenridge, D.A., Nealon, J.O. & Philomina, L.S. (2019) IntFOLD: an integrated web resource for high performance protein structure and function prediction. Nucleic Acids Research, 47, W408-W413. DOI PubMed
News and updates
- Nov 2025: Paper published in The Journal of Clinical Endocrinology & Metabolism - Publications
- Aug 2025: Our paper on the performance of ModFOLDdock2 at CASP16 has been published in the CASP special issue of Proteins - Publications
- Jun 2025: Docker package for FunFOLD5 is now available: https://hub.docker.com/r/radiyaman/funfold5_template
- Apr 2025: Paper describing the MultiFOLD2 and ModFOLDdock2 servers has been published in Nucleic Acids Research - Publications
- Mar 2025: Haematologica paper accepted - Publications
- Jan 2025: Docker package for MultiFOLD2, MultiFOLD2_refine and ModFOLDdock2 is now available: https://hub.docker.com/r/mcguffin/multifold2
- Dec 2024: Success at CASP16 and CAMEO: ModFOLDdock2 was ranked first in CASP16 for scoring global and local interfaces of quaternary structure models; MultiFOLD2 outperforms AF3 in CAMEO due to integrated stoichiometry prediction and was the top-ranked server on the hardest CASP16 domain targets according to GDT_TS.
- Oct 2024: New PhD student started - People
- Sep 2024: Nature Comms paper published - Publications
- Aug 2024: Bioinformatics paper published - Publications
- May-August 2022: CASP16 prediction season. New servers and methods have been developed, which will be independently benchmarked, including: MultiFOLD2, ModFOLDdock2 and FunFOLD5.
- Apr 2024: New PhD student started - People
- Mar 2024: JMB paper describing ModFOLD9 has been published - Publications
- Jan 2024: New server hardware has been installed.
- Dec 2023: The ModFOLD9 server is now available ModFOLD
- Jun 2023: Papers describing the ModFOLDdock and MultiFOLD_refine methods published in Proteins and Bioinformatics Advances, respectively - Publications
- Apr 2023: Paper describing the IntFOLD7, MultiFOLD and ModFOLDdock servers has been accepted by Nucleic Acids Research - Publications
- Mar 2023: Docker package for MultiFOLD, MultiFOLD_refine and ModFOLDdock is now available: https://hub.docker.com/r/mcguffin/multifold
- Dec 2022: Success at CASP and CAMEO: IntFOLD7 is competitive with AlphaFold2 baselines and other new Deep Learning methods in terms of tertiary structure prediction, while also integrating leading model quality estimates, protein-ligand interaction predictions, disorder and domain prediction.
- May 2022: CASP15 prediction season begins! New servers and methods have been developed, which will be independently benchmarked, including: IntFOLD7, FunFOLD4 and ModFOLD9.
- March 2022: new versions of our servers and methods have been registered with CAMEO and are being independently benchmarked, including: IntFOLD7, ModFOLD9 and ModFOLD9_pure.
- Oct 2021: New server hardware has been installed.
- Aug 2021: Proteins paper accepted - "Modeling SARS-CoV2 proteins in the CASP-commons experiment" - Publications
- Feb 2021: Two post docs are now working on our BBSRC project - People
- Jan 2021: New hardware installed for IntFOLD component methods.
- Jun 2020: Press release for CASP Commons research into SARS-CoV-2 protein structures.
- May 2020: CASP14 prediction season begins! New servers and methods will be benchmarked.
- Mar-Jun 2020: COVID-19 research - SARS-CoV-2 protein structures have been modelled using IntFOLD6 and scored with ModFOLD8 as part of CASP Commons 2020
- Feb 2020: New hardware has been installed for the new versions of our methods - IntFOLD6 and ModFOLD8 will be benchmarked in CASP14 and CAMEO.
- Apr 2019: IntFOLD5 server paper accepted by Nucleic Acids Research.
- Dec 2018: Success at CASP13 (we are among the top few ranked groups in QA and TBM)! Invited talk & panel contribution given and two posters presented on our new methods and servers. Quoted in Guardian article re: CASP13 and DeepMind
- Nov-Dec 2018: user interface improvements and new server hardware.
- May-Aug 2018: CASP13 prediction season.
- May 2018: IntFOLD5 server methods ready for CASP13
- Jul 2017: Template Based Modelling paper accepted for the CASP12 Proteins Special Issue
- Dec 2016: Success at CASP12 (among the top few ranked in TBM and QA)! Two talks and three posters presented on our new methods and servers. The ReFOLD and ModFOLD6 servers are now online.
- Feb 2015: Science paper published containing results from the IntFOLD server. Press release.
- Dec 2014: new interactive model visualisations implemented using the JSmol/HTML5 framework (also works on tablets and phones).
- Dec 2014: improved job status notification system implemented.
- Dec 2014: Success at CASP11, especially in the Quality Assessment (QA3) category
- Aug 2014: CASP11 prediction season complete.
- May 2014: CASP11 starts! New versions of the server component methods will be benchmarked. Provision of new CASP & CAMEO structural and functional data types/formats.
- Sep 2013: IntFOLD2 integrated with the Protein Model Portal
- Dec 2012: success at CASP10 - short talks given on quality assessment and function prediction
- Dec 2012: paper reporting extensive application of servers has been published in BMC Genomics
- Aug 2012: IntFOLD2 open beta version online for testing please send feedback to l.j.mcguffin@reading.ac.uk
- May 2012: Multi-Template Modelling (MTM) method Bioinformatics paper published (Buenavista et al., Bioinformatics, 2012), the method forms the basis of IntFOLD2-TS predictions
- Sept 2011: Confidence scores (p-values) now included in graphical output. Help page
- Aug 2011: Interactive IntFOLD-TS output now includes model-template superpositions - Example
- May 2011: IntFOLD-TS method paper now in press (CASP9 special issue) - Publications
- May 2011: FunFOLD paper now in press - Publications Download FunFOLD
- March 2011: IntFOLD server paper now in press (NAR Web Server issue) - Publications
- March 2011: Methods paper describing application of servers to Blumeria proteome now in press - Publications
- Dec 2010: The IntFOLD server version is now out of beta.
- Dec 2010: The IntFOLD-TS method was the focus of our invited talk for the CASP9 methods session. Download slides (.pptx).