The IntFOLD Server Results Page (Version 7.0)
Please cite the relevant paper:McGuffin, L. J., Edmunds N. S., Genc, A. G., Alharbi, S. M. A., Salehe, B. R. and Adiyaman, R. (2023) Prediction of protein structures, functions and interactions using the IntFOLD7, MultiFOLD and ModFOLDdock servers. Nucleic Acids Research. gkad297. DOI PubMed
Links to graphical output:
- Top 5 3D models
- Disorder prediction
- Domain boundary prediction
- Binding site prediction
- Full model quality assessment results
Download machine readable results in CASP format:
- TS (Tertiary Structure Prediction)
- DR (Disorder Prediction)
- DP (Domain Prediction)
- FN (Binding Site Prediction)
- QA (Model Quality Prediction)
- Make a compressed file containing all generated 3D models (models are in PDB format with predicted per-residue errors in the B-factor column)
Results will be available for 21 days after job completion (subject to server capacity)
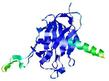
| Tertiary Structure Predictions: Top 5 multi-template 3D models for H1135_A | Help | ||||
| Model ID and PDBsum links for all templates used | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | 3D map of model quality |
|
IntFOLD7 AF2 TS3 4 0 |
CERT: 2.284E-5 |
0.7598 |
 |
|
|
IntFOLD7 AF2 TS3 4 0 rel |
CERT: 2.399E-5 |
0.7572 |
 |

|
|
IntFOLD7 wPPAS multi1 TS1 3unpA 5ywzA2 |
CERT: 2.449E-5 |
0.7561 |
 |

|
|
IntFOLD7 wdPPAS multi1 TS1 3unpA 5ywzA2 |
CERT: 2.449E-5 |
0.7561 |
 |

|
|
IntFOLD7 AF2 TS4 1 0 |
CERT: 2.51E-5 |
0.7548 |
 |

|
McGuffin, L. J. (2008) Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics, 24, 1789-1804. PubMed
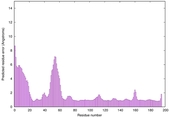
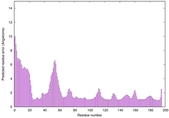
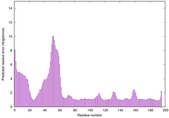
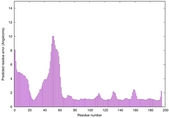
| Disorder prediction for H1135_A | Help | |||
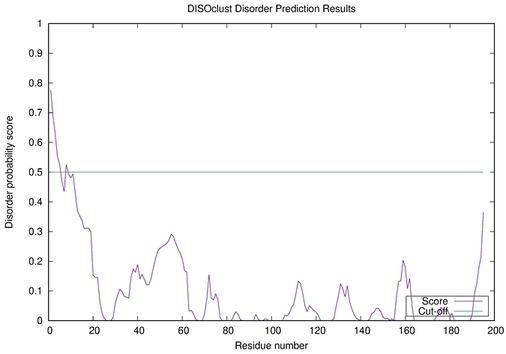
| |||
| Click image to download plot in PostScript format. |
| Domain boundary prediction for H1135_A | Help | |||
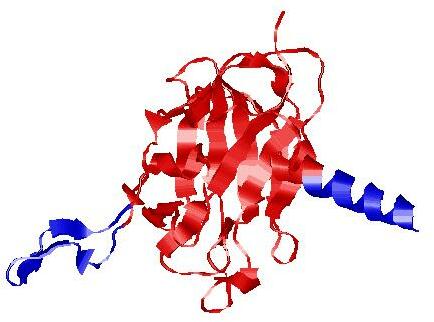
| |||
| Click image to download model or view prediction using Jmol. The model above is coloured according to the predicted domains. |
Roche, D. B., Buenavista, M. T., McGuffin, L. J. (2013) The FunFOLD2 server for the prediction of protein-ligand interactions. Nucleic Acids Res., 41, W303-7. PubMed
| Ligand binding residues prediction for H1135_A | Help | |||
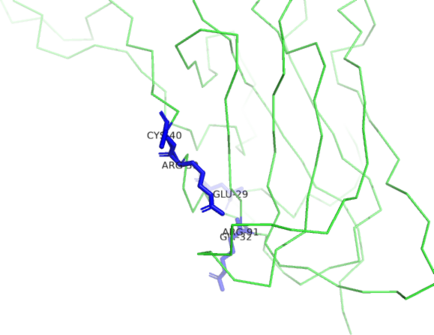
| |||
| Click image to download model or view prediction using Jmol. Predicted ligand binding residues are shown as blue sticks in the image above. |
McGuffin, L.J., Aldowsari, F., Alharbi, S. and Adiyaman, R. (2021) ModFOLD8: accurate global and local quality estimates for 3D protein models. Nucleic Acids Research, 49, W425-W430. DOI PubMed
| Full model quality assessment results for H1135_A | Help | ||||
| Model ID and PDBsum links for all templates used | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | 3D map of model quality |
|
IntFOLD7 AF2 TS3 4 0 |
CERT: 2.284E-5 |
0.7598 |
 |

|
|
IntFOLD7 AF2 TS3 4 0 rel |
CERT: 2.399E-5 |
0.7572 |
 |

|
|
IntFOLD7 wPPAS multi1 TS1 3unpA 5ywzA2 |
CERT: 2.449E-5 |
0.7561 |
 |

|
|
IntFOLD7 wdPPAS multi1 TS1 3unpA 5ywzA2 |
CERT: 2.449E-5 |
0.7561 |
 |

|
|
IntFOLD7 AF2 TS4 1 0 |
CERT: 2.51E-5 |
0.7548 |
 |

|
|
IntFOLD7 AF2 TS2 2 0 |
CERT: 2.513E-5 |
0.7547 |
 |

|
|
IntFOLD7 AF2 TS4 1 0 rel |
CERT: 2.755E-5 |
0.7498 |
 |

|
|
IntFOLD7 IntFOLDTS60 multi3 TS1 4dxtA 5ywzA |
CERT: 2.774E-5 |
0.7495 |
 |

|
|
IntFOLD7 AF2 TS5 3 0 rel |
CERT: 2.83E-5 |
0.7484 |
 |

|
|
IntFOLD7 AF2 TS2 2 0 rel |
CERT: 2.832E-5 |
0.7484 |
 |

|
|
IntFOLD7 dPPAS2 multi7 TS1 3unpA 5ywzA 5ywzA2 |
CERT: 2.848E-5 |
0.7481 |
 |

|
|
IntFOLD7 AF2 TS5 3 0 |
CERT: 2.856E-5 |
0.7479 |
 |

|
|
IntFOLD7 COMA multi8 TS1 5ywzA |
CERT: 2.858E-5 |
0.7479 |
 |

|
|
IntFOLD7 COMA multi7 TS1 5ywzA |
CERT: 2.858E-5 |
0.7479 |
 |

|
|
IntFOLD7 COMA multi6 TS1 5ywzA |
CERT: 2.858E-5 |
0.7479 |
 |

|
|
IntFOLD7 COMA multi4 TS1 5ywzA |
CERT: 2.858E-5 |
0.7479 |
 |

|
|
IntFOLD7 COMA multi3 TS1 5ywzA |
CERT: 2.858E-5 |
0.7479 |
 |

|
|
IntFOLD7 COMA multi2 TS1 5ywzA |
CERT: 2.858E-5 |
0.7479 |
 |

|
|
IntFOLD7 CNFsearch multi8 TS1 5ywzA |
CERT: 2.858E-5 |
0.7479 |
 |

|
|
IntFOLD7 CNFsearch multi7 TS1 5ywzA |
CERT: 2.858E-5 |
0.7479 |
 |

|
|
IntFOLD7 CNFsearch multi6 TS1 5ywzA |
CERT: 2.858E-5 |
0.7479 |
 |

|
|
IntFOLD7 CNFsearch multi4 TS1 5ywzA |
CERT: 2.858E-5 |
0.7479 |
 |

|
|
IntFOLD7 CNFsearch multi3 TS1 5ywzA |
CERT: 2.858E-5 |
0.7479 |
 |

|
|
IntFOLD7 CNFsearch multi2 TS1 5ywzA |
CERT: 2.858E-5 |
0.7479 |
 |

|
|
IntFOLD7 dPPAS2 multi5 TS1 3unpA 5ywzA |
CERT: 2.865E-5 |
0.7478 |
 |

|
|
IntFOLD7 wMUSTER multi5 TS1 5ywzA 5ywzA2 |
CERT: 2.869E-5 |
0.7477 |
 |

|
|
IntFOLD7 AF2 TS1 5 0 |
CERT: 2.895E-5 |
0.7472 |
 |

|
|
IntFOLD7 wPPAS multi7 TS1 5ywzA 5ywzA2 |
CERT: 3.011E-5 |
0.7451 |
 |

|
|
IntFOLD7 wPPAS multi5 TS1 5ywzA 5ywzA2 |
CERT: 3.011E-5 |
0.7451 |
 |

|
|
IntFOLD7 wdPPAS multi7 TS1 5ywzA 5ywzA2 |
CERT: 3.011E-5 |
0.7451 |
 |

|
|
IntFOLD7 wdPPAS multi5 TS1 5ywzA 5ywzA2 |
CERT: 3.011E-5 |
0.7451 |
 |

|
|
IntFOLD7 PPAS multi7 TS1 5ywzA 5ywzA2 |
CERT: 3.011E-5 |
0.7451 |
 |

|
|
IntFOLD7 PPAS multi5 TS1 5ywzA 5ywzA2 |
CERT: 3.011E-5 |
0.7451 |
 |

|
|
IntFOLD7 Env-PPAS multi7 TS1 5ywzA 5ywzA2 |
CERT: 3.011E-5 |
0.7451 |
 |

|
|
IntFOLD7 Env-PPAS multi5 TS1 5ywzA 5ywzA2 |
CERT: 3.011E-5 |
0.7451 |
 |

|
|
IntFOLD7 dPPAS multi7 TS1 5ywzA 5ywzA2 |
CERT: 3.011E-5 |
0.7451 |
 |

|
|
IntFOLD7 dPPAS multi5 TS1 5ywzA 5ywzA2 |
CERT: 3.011E-5 |
0.7451 |
 |

|
|
IntFOLD7 HHsearch multi8 TS1 5ywzA |
CERT: 3.111E-5 |
0.7434 |
 |

|
|
IntFOLD7 HHsearch multi7 TS1 5ywzA |
CERT: 3.111E-5 |
0.7434 |
 |

|
|
IntFOLD7 HHsearch multi6 TS1 5ywzA |
CERT: 3.111E-5 |
0.7434 |
 |

|
|
IntFOLD7 HHsearch multi4 TS1 5ywzA |
CERT: 3.111E-5 |
0.7434 |
 |

|
|
IntFOLD7 HHsearch multi3 TS1 5ywzA |
CERT: 3.111E-5 |
0.7434 |
 |

|
|
IntFOLD7 HHsearch multi2 TS1 5ywzA |
CERT: 3.111E-5 |
0.7434 |
 |

|
|
IntFOLD7 wPPAS multi8 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 wPPAS multi6 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 wdPPAS multi8 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 wdPPAS multi6 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 SPARKSX multi7 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 SPARKSX multi6 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 PPAS multi8 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 PPAS multi6 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 IntFOLDTS140 multi3 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 IntFOLDTS140 multi2 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 Env-PPAS multi8 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 Env-PPAS multi6 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 dPPAS multi8 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 dPPAS multi6 TS1 5ywzA |
CERT: 3.145E-5 |
0.7428 |
 |

|
|
IntFOLD7 IntFOLDTS140 multi1 TS1 5ywzA |
CERT: 3.216E-5 |
0.7416 |
 |

|
|
IntFOLD7 wMUSTER multi8 TS1 5ywzA |
CERT: 3.222E-5 |
0.7415 |
 |

|
|
IntFOLD7 wMUSTER multi7 TS1 5ywzA |
CERT: 3.222E-5 |
0.7415 |
 |

|
|
IntFOLD7 wMUSTER multi6 TS1 5ywzA |
CERT: 3.222E-5 |
0.7415 |
 |

|
|
IntFOLD7 wMUSTER multi4 TS1 5ywzA |
CERT: 3.222E-5 |
0.7415 |
 |

|
|
IntFOLD7 wMUSTER multi3 TS1 5ywzA |
CERT: 3.222E-5 |
0.7415 |
 |

|
|
IntFOLD7 wMUSTER multi2 TS1 5ywzA |
CERT: 3.222E-5 |
0.7415 |
 |

|
|
IntFOLD7 wMUSTER multi1 TS1 5ywzA 3unpA |
CERT: 3.354E-5 |
0.7394 |
 |

|
|
IntFOLD7 AF2 TS1 5 0 rel |
CERT: 3.507E-5 |
0.7370 |
 |

|
|
IntFOLD7 PPAS multi3 TS1 5ywzA2 |
CERT: 3.576E-5 |
0.7360 |
 |

|
|
IntFOLD7 PPAS multi2 TS1 5ywzA2 |
CERT: 3.576E-5 |
0.7360 |
 |

|
|
IntFOLD7 LOMETS multi3 TS1 5ywzA2 |
CERT: 3.576E-5 |
0.7360 |
 |

|
|
IntFOLD7 LOMETS multi2 TS1 5ywzA2 |
CERT: 3.576E-5 |
0.7360 |
 |

|
|
IntFOLD7 LOMETS multi1 TS1 5ywzA2 |
CERT: 3.576E-5 |
0.7360 |
 |

|
|
IntFOLD7 Env-PPAS multi3 TS1 5ywzA2 |
CERT: 3.576E-5 |
0.7360 |
 |

|
|
IntFOLD7 Env-PPAS multi2 TS1 5ywzA2 |
CERT: 3.576E-5 |
0.7360 |
 |

|
|
IntFOLD7 dPPAS multi3 TS1 5ywzA2 |
CERT: 3.576E-5 |
0.7360 |
 |

|
|
IntFOLD7 dPPAS multi2 TS1 5ywzA2 |
CERT: 3.576E-5 |
0.7360 |
 |

|
|
IntFOLD7 dPPAS2 multi3 TS1 5ywzA2 |
CERT: 3.576E-5 |
0.7360 |
 |

|
|
IntFOLD7 dPPAS2 multi2 TS1 5ywzA2 |
CERT: 3.576E-5 |
0.7360 |
 |

|
|
IntFOLD7 COMA multi5 TS1 5ywzA 3unpA |
CERT: 3.602E-5 |
0.7356 |
 |

|
|
IntFOLD7 COMA multi1 TS1 5ywzA 3unpA |
CERT: 3.602E-5 |
0.7356 |
 |

|
|
IntFOLD7 wPPAS multi3 TS1 3unpA 5ywzA2 5ywzA |
CERT: 3.951E-5 |
0.7306 |
 |

|
|
IntFOLD7 wdPPAS multi3 TS1 3unpA 5ywzA2 5ywzA |
CERT: 3.951E-5 |
0.7306 |
 |

|
|
IntFOLD7 PPAS multi4 TS1 5ywzA2 3unpA |
CERT: 4.093E-5 |
0.7288 |
 |

|
|
IntFOLD7 PPAS multi1 TS1 5ywzA2 3unpA |
CERT: 4.093E-5 |
0.7288 |
 |

|
|
IntFOLD7 dPPAS multi4 TS1 5ywzA2 3unpA |
CERT: 4.093E-5 |
0.7288 |
 |

|
|
IntFOLD7 dPPAS multi1 TS1 5ywzA2 3unpA |
CERT: 4.093E-5 |
0.7288 |
 |

|
|
IntFOLD7 dPPAS2 multi4 TS1 5ywzA2 3unpA |
CERT: 4.093E-5 |
0.7288 |
 |

|
|
IntFOLD7 dPPAS2 multi1 TS1 5ywzA2 3unpA |
CERT: 4.093E-5 |
0.7288 |
 |

|
|
IntFOLD7 LOMETS multi4 TS1 5ywzA2 3unpA |
CERT: 4.101E-5 |
0.7287 |
 |

|
|
IntFOLD7 Env-PPAS multi4 TS1 5ywzA2 3unpA |
CERT: 4.101E-5 |
0.7287 |
 |

|
|
IntFOLD7 Env-PPAS multi1 TS1 5ywzA2 3unpA |
CERT: 4.101E-5 |
0.7287 |
 |

|
|
IntFOLD7 CNFsearch multi5 TS1 5ywzA 5ed8A |
CERT: 4.265E-5 |
0.7266 |
 |

|
|
IntFOLD7 CNFsearch multi1 TS1 5ywzA 5ed8A |
CERT: 4.265E-5 |
0.7266 |
 |

|
|
IntFOLD7 HHsearch multi5 TS1 5ywzA 4dxtA |
CERT: 4.513E-5 |
0.7236 |
 |

|
|
IntFOLD7 HHsearch multi1 TS1 5ywzA 4dxtA |
CERT: 4.513E-5 |
0.7236 |
 |

|
|
IntFOLD7 SPARKSX multi5 TS1 5ywzA 3unpA |
CERT: 4.767E-5 |
0.7207 |
 |

|
|
IntFOLD7 SPARKSX multi2 TS1 6r15A |
CERT: 6.272E-5 |
0.7060 |
 |

|
|
IntFOLD7 IT5 TS3 |
CERT: 6.855E-5 |
0.7013 |
 |

|
|
IntFOLD7 SPARKSX multi3 TS1 6r15A 5ywzA |
CERT: 8.436E-5 |
0.6903 |
 |

|
|
IntFOLD7 SPARKSX multi1 TS1 6r15A 5ywzA |
CERT: 8.436E-5 |
0.6903 |
 |

|
|
IntFOLD7 HHpred TS1 4dxtA 5ed8A |
CERT: 1.104E-4 |
0.6760 |
 |

|
|
IntFOLD7 HHpred TS2 4dxtA 5ed8A |
CERT: 1.134E-4 |
0.6745 |
 |

|
|
IntFOLD7 IT5 TS1 |
CERT: 1.168E-4 |
0.6730 |
 |

|
|
IntFOLD7 IntFOLDTS60 multi1 TS1 4dxtA 3unpA |
CERT: 1.244E-4 |
0.6696 |
 |

|
|
IntFOLD7 IntFOLDTS60 multi2 TS1 4dxtA |
CERT: 1.383E-4 |
0.6640 |
 |

|
|
IntFOLD7 HHpred TS3 4dxtA 5ed8A |
CERT: 2.224E-4 |
0.6387 |
 |

|
|
IntFOLD7 tR2 TS0 1 0.25 |
CERT: 2.557E-4 |
0.6313 |
 |

|
|
IntFOLD7 tR2 TS0 2 0.15 |
CERT: 2.621E-4 |
0.6299 |
 |

|
|
IntFOLD7 tR2 TS0 1 0.45 |
CERT: 3.077E-4 |
0.6214 |
 |

|
|
IntFOLD7 tR2 TS0 0 0.05 |
CERT: 3.145E-4 |
0.6202 |
 |

|
|
IntFOLD7 tR2 TS0 0 0.15 |
CERT: 3.263E-4 |
0.6183 |
 |

|
|
IntFOLD7 tR2 TS0 0 0.35 |
CERT: 3.284E-4 |
0.6179 |
 |

|
|
IntFOLD7 dPPAS2 multi8 TS1 3unpA |
CERT: 3.389E-4 |
0.6163 |
 |

|
|
IntFOLD7 dPPAS2 multi6 TS1 3unpA |
CERT: 3.389E-4 |
0.6163 |
 |

|
|
IntFOLD7 wPPAS multi4 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 wPPAS multi2 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 wdPPAS multi4 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 wdPPAS multi2 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 spk2 multi8 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 spk2 multi7 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 spk2 multi6 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 spk2 multi4 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 spk2 multi3 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 spk2 multi2 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 sp3 multi8 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 sp3 multi7 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 sp3 multi6 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 sp3 multi4 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 sp3 multi3 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 sp3 multi2 TS1 3unpA |
CERT: 3.688E-4 |
0.6118 |
 |

|
|
IntFOLD7 tR2 TS0 2 0.45 |
CERT: 3.783E-4 |
0.6104 |
 |

|
|
IntFOLD7 tR2 TS0 2 0.25 |
CERT: 3.789E-4 |
0.6103 |
 |

|
|
IntFOLD7 tR2 TS0 0 0.25 |
CERT: 3.854E-4 |
0.6094 |
 |

|
|
IntFOLD7 tR2 TS0 1 0.35 |
CERT: 3.902E-4 |
0.6088 |
 |

|
|
IntFOLD7 tR2 TS0 1 0.05 |
CERT: 4.032E-4 |
0.6070 |
 |

|
|
IntFOLD7 sp3 multi5 TS1 3unpA 7pwfH |
CERT: 4.117E-4 |
0.6059 |
 |

|
|
IntFOLD7 tR2 TS0 2 0.35 |
CERT: 4.299E-4 |
0.6036 |
 |

|
|
IntFOLD7 tR2 TS0 2 0.05 |
CERT: 4.373E-4 |
0.6027 |
 |

|
|
IntFOLD7 sp3 multi1 TS1 3unpA 7yxvA |
CERT: 4.46E-4 |
0.6016 |
 |

|
|
IntFOLD7 tR2 TS0 1 0.15 |
CERT: 4.682E-4 |
0.5991 |
 |

|
|
IntFOLD7 tR2 TS0 0 0.45 |
CERT: 5.392E-4 |
0.5915 |
 |

|
|
IntFOLD7 spk2 multi5 TS1 3unpA 7pwfB |
CERT: 6.248E-4 |
0.5837 |
 |

|
|
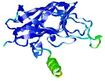
IntFOLD7 IT5 TS2 |
CERT: 7.575E-4 |
0.5735 |
 |
|
|
IntFOLD7 spk2 multi1 TS1 3unpA 7u7nD |
HIGH: 2.329E-3 |
0.5137 |
 |

|
|
IntFOLD7 DMPf TS1 |
HIGH: 4.335E-3 |
0.4806 |
 |

|
|
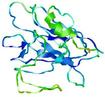
IntFOLD7 DMPf TS2 |
HIGH: 6.471E-3 |
0.4593 |
 |
|