ModFOLDdock: quality estimates for protein quaternary structure models
About the server
The ModFOLDdock server provides:
- World leading estimates of model accuracy for quaternary structures of proteins.
- Global assembly and interface quality scores, plus local per-residue interface scores
- Interactive 3D views of multimeric assemblies ranked by predicted model quality, which can be coloured by chain ID or interface accuracy.
- Machine-readable data downloads.
This website is free and open to all users and there is no login requirement.
Privacy and cookies
Fair usage policy:
You are only permitted to have 1 job running at a time for each IP address, so please wait until your previous job completes before submitting further data. If you already have a job running then you will be notified and your uploaded data will be deleted. Once your job has completed your IP address will be unlocked and you will be able to submit new data.
New! ModFOLDdock2 server submission form
- ModFOLDdock2 is the best method according to the CASP16 assessors for predicting both the global (QSCORE) and local interface accuracy of modelled protein complexes.
- The ModFOLDdock2 variants ranked within the top few groups at CASP16 in all QMODE1/2 metrics.
- We were invited to talk about our ModFOLDdock2 methods at the CASP16 conference in December 2024.
ModFOLDdock server submission form
Help page
Sample output
References
The ModFOLDdock method is described in these papers:- Fadini, A., Adiyaman, R., Alhaddad, S. N., Behzadi, B., Cheng, J., Cui, X., Edmunds, N. S., Freddolino, L., Genc, A. G., Liang, F., Liu, D., Liu, J., Liu, Q., McGuffin, L. J., Neupane, P., Peng, C., Shortle, D. R., Sun, M., Wang, H., Wuyun, Q., Zhang, G., Zhao, X., Zheng, W., & Read, R. J. (2025) Highlights of Model Quality Assessment in CASP16. Proteins, 94, 314-329. DOI PubMed
- Edmunds, N. S., Alharbi, S. M. A., Genc, A. G., Adiyaman, R. and McGuffin, L. J. (2023) Estimation of Model Accuracy in CASP15 Using the ModFOLDdock Server. Proteins, 91, 1871-1878. DOI PubMed
- McGuffin, L. J., Alhaddad, S. N., Behzadi, B., Edmunds, N. S., Genc, A. G., and Adiyaman, R. (2025) Prediction and quality assessment of protein quaternary structure models using the MultiFOLD2 and ModFOLDdock2 servers. Nucleic Acids Research, gkaf336. DOI PubMed
- McGuffin, L. J., Edmunds N. S., Genc, A. G., Alharbi, S. M. A., Salehe, B. R. and Adiyaman, R. (2023) Prediction of protein structures, functions and interactions using the IntFOLD7, MultiFOLD and ModFOLDdock servers. Nucleic Acids Research. gkad297. DOI PubMed
Docker package
- The Docker package for MultiFOLD2, MultiFOLD2_refine and ModFOLDdock2 is available here: https://hub.docker.com/r/mcguffin/multifold2
- The Docker package for MultiFOLD, MultiFOLD_refine and ModFOLDdock is available here: https://hub.docker.com/r/mcguffin/multifold

Predicted assembly quality scores versus oligo-lDDT scores for homomeric CASP15 targets
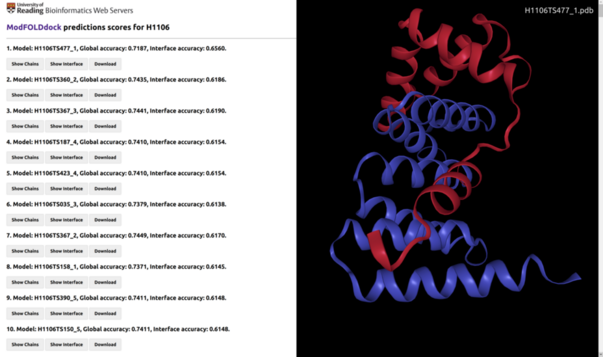
Example of the desktop view of the ModFOLDdock results page for CASP target H1106
Example of the mobile (portrait) view of the ModFOLDdock results page for CASP target H1106