The ModFOLD Server Results
Please cite the following papers:Maghrabi, A.H.A. & McGuffin, L.J. (2017) ModFOLD6: an accurate web server for the global and local quality estimation of 3D models of proteins, 45, W416-W421, Nucleic Acids Res. doi: 10.1093/nar/gkx332. PubMed
McGuffin, L.J., Shuid, A.M., Kempster, R., Maghrabi, A.H.A., Nealon J.O., Salehe, B.R., Atkins, J.D. & Roche, D.B. (2017) Accurate Template Based Modelling in CASP12 using the IntFOLD4-TS, ModFOLD6 and ReFOLD methods. Proteins: Structure, Function, and Bioinformatics, 86 Suppl 1, 335-344, doi: 10.1002/prot.25360. PubMed
Click on the links below to download machine readable results files:
- Download Quality Assessment file (raw model quality prediction scores in CASP QA format)
- Make a compressed file containing all evaluated 3D models (models are in PDB format with predicted per-residue errors in the B-factor column)
Results will be available for 21 days after job completion (subject to server capacity)
| Graphical ModFOLD6 results for T0862 | Help | ||||
| Model name | Confidence and P-value | Global model quality score | Residue error plot (click image for large version) | 3D view of residue error (click image for large version) |
| QUARK_TS3 | MEDIUM: 1.457E-2 |
0.3807 |
 |
 |
| Zhang-Server _TS1 |
MEDIUM: 3.129E-2 |
0.3634 |
 |
 |
| GOAL_TS1 | MEDIUM: 3.234E-2 |
0.3625 |
 |
 |
| Zhang-Server _TS2 |
MEDIUM: 3.307E-2 |
0.3620 |
 |
 |
| BAKER-ROSETT ASERVER_TS2 |
MEDIUM: 3.543E-2 |
0.3601 |
 |
 |
| Zhang-Server _TS3 |
MEDIUM: 3.772E-2 |
0.3583 |
 |
 |
| Zhang-Server _TS4 |
MEDIUM: 3.904E-2 |
0.3573 |
 |
 |
| RBO_Aleph_TS 3 |
MEDIUM: 3.938E-2 |
0.3571 |
 |
 |
| RBO_Aleph_TS 5 |
MEDIUM: 4.051E-2 |
0.3563 |
 |
 |
| BAKER-ROSETT ASERVER_TS1 |
MEDIUM: 4.079E-2 |
0.3561 |
 |
 |
| QUARK_TS2 | MEDIUM: 4.155E-2 |
0.3555 |
 |
 |
| QUARK_TS4 | MEDIUM: 4.175E-2 |
0.3554 |
 |
 |
| Zhang-Server _TS5 |
MEDIUM: 4.216E-2 |
0.3551 |
 |
 |
| RBO_Aleph_TS 2 |
MEDIUM: 4.699E-2 |
0.3517 |
 |
 |
| BAKER-ROSETT ASERVER_TS5 |
MEDIUM: 4.915E-2 |
0.3503 |
 |
 |
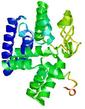
| QUARK_TS5 | MEDIUM: 4.953E-2 |
0.3501 |
 |
 |
| BAKER-ROSETT ASERVER_TS3 |
LOW: 5.215E-2 |
0.3484 |
 |
 |
| QUARK_TS1 | LOW: 5.233E-2 |
0.3482 |
 |
 |
| RBO_Aleph_TS 1 |
LOW: 5.37E-2 |
0.3474 |
 |
 |
| RBO_Aleph_TS 4 |
LOW: 5.963E-2 |
0.3438 |
 |
 |
| RaptorX_TS1 | LOW: 6.447E-2 |
0.3410 |
 |
 |
| MULTICOM-CON STRUCT_TS1 |
LOW: 6.524E-2 |
0.3406 |
 |
 |
| MULTICOM-CLU STER_TS2 |
LOW: 6.524E-2 |
0.3406 |
 |
 |
| MULTICOM-CON STRUCT_TS3 |
LOW: 7.059E-2 |
0.3377 |
 |
 |
| MULTICOM-CLU STER_TS3 |
LOW: 7.059E-2 |
0.3377 |
 |
 |
| MULTICOM-NOV EL_TS2 |
LOW: 7.112E-2 |
0.3374 |
 |
 |
| MULTICOM-NOV EL_TS1 |
LOW: 7.27E-2 |
0.3366 |
 |
 |
| MULTICOM-REF INE_TS1 |
LOW: 7.518E-2 |
0.3353 |
 |
 |
| MULTICOM-CON STRUCT_TS5 |
LOW: 7.78E-2 |
0.3340 |
 |
 |
| MULTICOM-CLU STER_TS5 |
LOW: 7.78E-2 |
0.3340 |
 |
 |
| MULTICOM-REF INE_TS3 |
LOW: 8.071E-2 |
0.3326 |
 |
 |
| MULTICOM-NOV EL_TS3 |
LOW: 8.081E-2 |
0.3326 |
 |
 |
| PhyreTopoAlp ha_TS5 |
LOW: 8.45E-2 |
0.3308 |
 |
 |
| GOAL_TS2 | LOW: 8.521E-2 |
0.3305 |
 |
 |
| MULTICOM-REF INE_TS5 |
LOW: 9.074E-2 |
0.3281 |
 |
 |
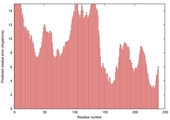
| Pcons-net_TS 1 |
LOW: 9.558E-2 |
0.3260 |
 |
 |
| Pcons-net_TS 2 |
LOW: 9.734E-2 |
0.3253 |
 |
 |
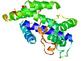
| BAKER-ROSETT ASERVER_TS4 |
LOW: 9.802E-2 |
0.3250 |
 |
 |
| Pcons-net_TS 4 |
LOW: 9.996E-2 |
0.3243 |
 |
 |
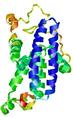
| MULTICOM-CLU STER_TS1 |
POOR: 1.005E-1 |
0.3240 |
 |
 |
| FALCON_TOPOX _TS3 |
POOR: 1.028E-1 |
0.3232 |
 |
 |
| Pareto-serve r_TS2 |
POOR: 1.069E-1 |
0.3216 |
 |
 |
| chuo-u-serve r_TS5 |
POOR: 1.086E-1 |
0.3210 |
 |
 |
| chuo-u2_TS5 | POOR: 1.086E-1 |
0.3210 |
 |
 |
| GAPF_LNCC_SE RVER_TS2 |
POOR: 1.095E-1 |
0.3206 |
 |
 |
| BhageerathH- Plus_TS2 |
POOR: 1.136E-1 |
0.3192 |
 |
 |
| YASARA_TS4 | POOR: 1.143E-1 |
0.3189 |
 |
 |
| ToyPred_emai l_TS1 |
POOR: 1.146E-1 |
0.3188 |
 |
 |
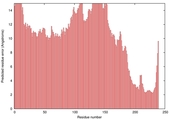
| MULTICOM-REF INE_TS2 |
POOR: 1.154E-1 |
0.3186 |
 |
 |
| MULTICOM-CON STRUCT_TS4 |
POOR: 1.173E-1 |
0.3179 |
 |
 |
| Pareto-serve r_TS1 |
POOR: 1.173E-1 |
0.3179 |
 |
 |
| MULTICOM-REF INE_TS4 |
POOR: 1.187E-1 |
0.3174 |
 |
 |
| FALCON_TOPO_ TS1 |
POOR: 1.189E-1 |
0.3174 |
 |
 |
| GOAL_TS3 | POOR: 1.19E-1 |
0.3173 |
 |
 |
| BhageerathH- Plus_TS1 |
POOR: 1.193E-1 |
0.3172 |
 |
 |
| GAPF_LNCC_SE RVER_TS3 |
POOR: 1.204E-1 |
0.3169 |
 |
 |
| FALCON_TOPOX _TS4 |
POOR: 1.248E-1 |
0.3154 |
 |
 |
| GOAL_TS4 | POOR: 1.26E-1 |
0.3150 |
 |
 |
| BhageerathH- Plus_TS3 |
POOR: 1.274E-1 |
0.3146 |
 |
 |
| FALCON_TOPOX _TS1 |
POOR: 1.296E-1 |
0.3139 |
 |
 |
| FALCON_TOPOX _TS5 |
POOR: 1.302E-1 |
0.3137 |
 |
 |
| Pcons-net_TS 5 |
POOR: 1.324E-1 |
0.3130 |
 |
 |
| PhyreTopoAlp ha_TS4 |
POOR: 1.363E-1 |
0.3119 |
 |
 |
| FALCON_TOPOX _TS2 |
POOR: 1.38E-1 |
0.3114 |
 |
 |
| Pcons-net_TS 3 |
POOR: 1.386E-1 |
0.3112 |
 |
 |
| FALCON_TOPO_ TS5 |
POOR: 1.393E-1 |
0.3110 |
 |
 |
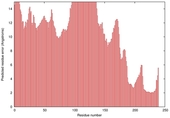
| BhageerathH- Plus_TS5 |
POOR: 1.425E-1 |
0.3101 |
 |
 |
| FALCON_TOPO_ TS4 |
POOR: 1.453E-1 |
0.3093 |
 |
 |
| Pareto-serve r_TS3 |
POOR: 1.548E-1 |
0.3067 |
 |
 |
| BhageerathH- Plus_TS4 |
POOR: 1.61E-1 |
0.3051 |
 |
 |
| FALCON_TOPO_ TS3 |
POOR: 1.613E-1 |
0.3051 |
 |
 |
| IntFOLD4_TS1 |
POOR: 1.678E-1 |
0.3034 |
 |
 |
| GAPF_LNCC_SE RVER_TS1 |
POOR: 1.726E-1 |
0.3023 |
 |
 |
| FALCON_TOPO_ TS2 |
POOR: 1.774E-1 |
0.3012 |
 |
 |
| Pareto-serve r_TS4 |
POOR: 1.831E-1 |
0.2999 |
 |
 |
| chuo-u-serve r_TS3 |
POOR: 1.84E-1 |
0.2996 |
 |
 |
| chuo-u2_TS3 | POOR: 1.84E-1 |
0.2996 |
 |
 |
| IntFOLD4_TS5 |
POOR: 1.847E-1 |
0.2995 |
 |
 |
| MUfold1_TS2 | POOR: 1.881E-1 |
0.2987 |
 |
 |
| GAPF_LNCC_SE RVER_TS4 |
POOR: 1.891E-1 |
0.2985 |
 |
 |
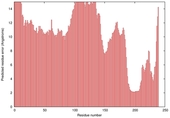
| Pareto-serve r_TS5 |
POOR: 1.949E-1 |
0.2973 |
 |
 |
| MUfold1_TS4 | POOR: 1.974E-1 |
0.2967 |
 |
 |
| GOAL_TS5 | POOR: 1.989E-1 |
0.2964 |
 |
 |
| PhyreTopoAlp ha_TS1 |
POOR: 2.052E-1 |
0.2951 |
 |
 |
| MUfold1_TS3 | POOR: 2.065E-1 |
0.2948 |
 |
 |
| IntFOLD4_TS4 |
POOR: 2.127E-1 |
0.2936 |
 |
 |
| MUfold1_TS5 | POOR: 2.295E-1 |
0.2904 |
 |
 |
| chuo-u-serve r_TS4 |
POOR: 2.334E-1 |
0.2896 |
 |
 |
| chuo-u2_TS4 | POOR: 2.334E-1 |
0.2896 |
 |
 |
| MUfold1_TS1 | POOR: 2.342E-1 |
0.2895 |
 |
 |
| chuo-u-serve r_TS1 |
POOR: 2.378E-1 |
0.2888 |
 |
 |
| chuo-u2_TS1 | POOR: 2.378E-1 |
0.2888 |
 |
 |
| PhyreTopoAlp ha_TS3 |
POOR: 2.413E-1 |
0.2882 |
 |
 |
| YASARA_TS2 | POOR: 2.425E-1 |
0.2880 |
 |
 |
| RaptorX-Cont act_TS4 |
POOR: 2.489E-1 |
0.2868 |
 |
 |
| YASARA_TS1 | POOR: 2.589E-1 |
0.2851 |
 |
 |
| RaptorX-Cont act_TS2 |
POOR: 2.662E-1 |
0.2839 |
 |
 |
| RaptorX-Cont act_TS1 |
POOR: 2.814E-1 |
0.2814 |
 |
 |
| slbio_TS1 | POOR: 2.923E-1 |
0.2796 |
 |
 |
| slbio_TS2 | POOR: 2.937E-1 |
0.2794 |
 |
 |
| slbio_TS4 | POOR: 2.999E-1 |
0.2784 |
 |
 |
| RaptorX-Cont act_TS5 |
POOR: 3.016E-1 |
0.2782 |
 |
 |
| GAPF_LNCC_SE RVER_TS5 |
POOR: 3.045E-1 |
0.2777 |
 |
 |
| slbio_TS3 | POOR: 3.192E-1 |
0.2755 |
 |
 |
| MULTICOM-NOV EL_TS4 |
POOR: 3.316E-1 |
0.2737 |
 |
 |
| MULTICOM-NOV EL_TS5 |
POOR: 3.33E-1 |
0.2735 |
 |
 |
| tsspred2_TS4 |
POOR: 3.381E-1 |
0.2727 |
 |
 |
| FFAS-3D_TS1 | POOR: 3.484E-1 |
0.2713 |
 |
 |
| PhyreTopoAlp ha_TS2 |
POOR: 3.492E-1 |
0.2711 |
 |
 |
| RaptorX-Cont act_TS3 |
POOR: 3.544E-1 |
0.2704 |
 |
 |
| YASARA_TS3 | POOR: 3.628E-1 |
0.2692 |
 |
 |
| tsspred2_TS5 |
POOR: 3.638E-1 |
0.2691 |
 |
 |
| myprotein-me _TS1 |
POOR: 3.659E-1 |
0.2688 |
 |
 |
| myprotein-me _TS3 |
POOR: 3.723E-1 |
0.2679 |
 |
 |
| Distill_TS3 | POOR: 3.74E-1 |
0.2677 |
 |
 |
| myprotein-me _TS5 |
POOR: 3.803E-1 |
0.2668 |
 |
 |
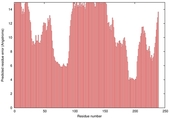
| myprotein-me _TS2 |
POOR: 4.001E-1 |
0.2641 |
 |
 |
| YASARA_TS5 | POOR: 4.076E-1 |
0.2631 |
 |
 |
| tsspred2_TS1 |
POOR: 4.42E-1 |
0.2586 |
 |
 |
| Distill_TS4 | POOR: 4.435E-1 |
0.2585 |
 |
 |
| tsspred2_TS3 |
POOR: 4.466E-1 |
0.2581 |
 |
 |
| slbio_TS5 | POOR: 4.539E-1 |
0.2571 |
 |
 |
| HHGG_TS1 | POOR: 4.77E-1 |
0.2542 |
 |
 |
| Atome2_CBS_T S5 |
POOR: 4.822E-1 |
0.2535 |
 |
 |
| myprotein-me _TS4 |
POOR: 4.864E-1 |
0.2530 |
 |
 |
| HHGG_TS4 | POOR: 4.902E-1 |
0.2525 |
 |
 |
| Seok-naive_a ssembly_TS4 |
POOR: 4.916E-1 |
0.2523 |
 |
 |
| Distill_TS2 | POOR: 5.273E-1 |
0.2478 |
 |
 |
| HHGG_TS3 | POOR: 5.277E-1 |
0.2478 |
 |
 |
| Seok-server_ TS1 |
POOR: 5.293E-1 |
0.2476 |
 |
 |
| HHGG_TS2 | POOR: 5.383E-1 |
0.2465 |
 |
 |
| Distill_TS1 | POOR: 5.443E-1 |
0.2457 |
 |
 |
| HHGG_TS5 | POOR: 5.525E-1 |
0.2447 |
 |
 |
| Atome2_CBS_T S3 |
POOR: 5.548E-1 |
0.2444 |
 |
 |
| Seok-server_ TS5 |
POOR: 5.631E-1 |
0.2433 |
 |
 |
| MUfold2_TS4 | POOR: 5.648E-1 |
0.2431 |
 |
 |
| Distill_TS5 | POOR: 5.776E-1 |
0.2415 |
 |
 |
| HHPred0_TS1 | POOR: 5.936E-1 |
0.2395 |
 |
 |
| Seok-server_ TS4 |
POOR: 5.973E-1 |
0.2390 |
 |
 |
| tsspred2_TS2 |
POOR: 6.14E-1 |
0.2369 |
 |
 |
| Seok-server_ TS2 |
POOR: 6.164E-1 |
0.2365 |
 |
 |
| HHPred1_TS1 | POOR: 6.343E-1 |
0.2342 |
 |
 |
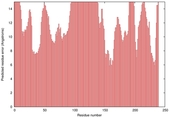
| Seok-server_ TS3 |
POOR: 6.467E-1 |
0.2326 |
 |
 |
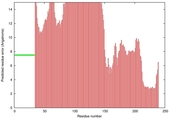
| MUfold2_TS3 | POOR: 6.672E-1 |
0.2298 |
 |
 |
| FLOUDAS_SERV ER_TS2 |
POOR: 7.841E-1 |
0.2124 |
 |
 |
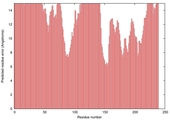
| Seok-assembl y_TS1 |
POOR: 7.995E-1 |
0.2097 |
 |
 |
| FLOUDAS_SERV ER_TS1 |
POOR: 8.028E-1 |
0.2091 |
 |
 |
| FLOUDAS_SERV ER_TS4 |
POOR: 8.248E-1 |
0.2050 |
 |
 |
| FLOUDAS_SERV ER_TS5 |
POOR: 8.277E-1 |
0.2044 |
 |
 |
| FLOUDAS_SERV ER_TS3 |
POOR: 8.479E-1 |
0.2002 |
 |
 |