The ModFOLD Server Results
Please cite the following papers:Maghrabi, A.H.A. & McGuffin, L.J. (2017) ModFOLD6: an accurate web server for the global and local quality estimation of 3D models of proteins, 45, W416-W421, Nucleic Acids Res. doi: 10.1093/nar/gkx332. PubMed
McGuffin, L.J., Shuid, A.M., Kempster, R., Maghrabi, A.H.A., Nealon J.O., Salehe, B.R., Atkins, J.D. & Roche, D.B. (2017) Accurate Template Based Modelling in CASP12 using the IntFOLD4-TS, ModFOLD6 and ReFOLD methods. Proteins: Structure, Function, and Bioinformatics, 86 Suppl 1, 335-344, doi: 10.1002/prot.25360. PubMed
Click on the links below to download machine readable results files:
- Download Quality Assessment file (raw model quality prediction scores in CASP QA format)
- Make a compressed file containing all evaluated 3D models (models are in PDB format with predicted per-residue errors in the B-factor column)
Results will be available for 21 days after job completion (subject to server capacity)
| Graphical ModFOLD6 results for T0860 | Help | ||||
| Model name | Confidence and P-value | Global model quality score | Residue error plot (click image for large version) | 3D view of residue error (click image for large version) |
| BAKER-ROSETT ASERVER_TS5 |
CERT: 1.229E-6 |
0.5984 |
 |
 |
| BAKER-ROSETT ASERVER_TS1 |
CERT: 1.244E-6 |
0.5981 |
 |
 |
| GOAL_COMPLEX _TS1 |
CERT: 3.049E-6 |
0.5773 |
 |
 |
| BAKER-ROSETT ASERVER_TS2 |
CERT: 3.084E-6 |
0.5771 |
 |
 |
| GOAL_COMPLEX _TS2 |
CERT: 3.186E-6 |
0.5763 |
 |
 |
| GOAL_TS1 | CERT: 3.628E-6 |
0.5733 |
 |
 |
| HHGG_TS4 | CERT: 4.001E-6 |
0.5711 |
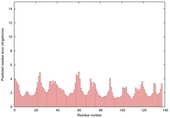 |
 |
| HHGG_TS5 | CERT: 4.121E-6 |
0.5704 |
 |
 |
| GOAL_COMPLEX _TS5 |
CERT: 4.285E-6 |
0.5695 |
 |
 |
| GOAL_TS3 | CERT: 4.524E-6 |
0.5682 |
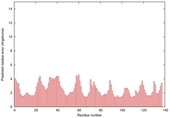 |
 |
| GOAL_TS4 | CERT: 4.822E-6 |
0.5667 |
 |
 |
| HHGG_TS1 | CERT: 4.933E-6 |
0.5662 |
 |
 |
| BAKER-ROSETT ASERVER_TS3 |
CERT: 5.218E-6 |
0.5649 |
 |
 |
| BAKER-ROSETT ASERVER_TS4 |
CERT: 5.234E-6 |
0.5648 |
 |
 |
| HHGG_TS3 | CERT: 5.248E-6 |
0.5648 |
 |
 |
| HHGG_TS2 | CERT: 5.319E-6 |
0.5645 |
 |
 |
| GOAL_COMPLEX _TS3 |
CERT: 5.502E-6 |
0.5637 |
 |
 |
| GOAL_COMPLEX _TS4 |
CERT: 5.797E-6 |
0.5625 |
 |
 |
| GOAL_TS2 | CERT: 6.486E-6 |
0.5599 |
 |
 |
| FALCON_TOPO_ TS1 |
CERT: 7.211E-6 |
0.5574 |
 |
 |
| Zhang-Server _TS2 |
CERT: 8.844E-6 |
0.5527 |
 |
 |
| GOAL_TS5 | CERT: 8.979E-6 |
0.5524 |
 |
 |
| FALCON_TOPOX _TS5 |
CERT: 9.94E-6 |
0.5500 |
 |
 |
| Atome2_CBS_T S3 |
CERT: 1.034E-5 |
0.5491 |
 |
 |
| Atome2_CBS_T S2 |
CERT: 1.056E-5 |
0.5486 |
 |
 |
| FALCON_TOPO_ TS4 |
CERT: 1.068E-5 |
0.5484 |
 |
 |
| Zhang-Server _TS1 |
CERT: 1.159E-5 |
0.5464 |
 |
 |
| FALCON_TOPOX _TS4 |
CERT: 1.176E-5 |
0.5461 |
 |
 |
| slbio_TS3 | CERT: 1.182E-5 |
0.5460 |
 |
 |
| slbio_TS5 | CERT: 1.199E-5 |
0.5457 |
 |
 |
| slbio_TS4 | CERT: 1.201E-5 |
0.5456 |
 |
 |
| Atome2_CBS_T S1 |
CERT: 1.228E-5 |
0.5451 |
 |
 |
| FALCON_TOPOX _TS2 |
CERT: 1.25E-5 |
0.5447 |
 |
 |
| FALCON_TOPO_ TS5 |
CERT: 1.342E-5 |
0.5431 |
 |
 |
| QUARK_TS2 | CERT: 1.372E-5 |
0.5425 |
 |
 |
| FALCON_TOPO_ TS3 |
CERT: 1.392E-5 |
0.5422 |
 |
 |
| HHPred0_TS1 | CERT: 1.441E-5 |
0.5414 |
 |
 |
| slbio_TS1 | CERT: 1.464E-5 |
0.5411 |
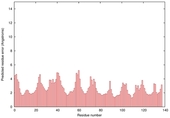 |
 |
| FALCON_TOPOX _TS3 |
CERT: 1.568E-5 |
0.5395 |
 |
 |
| FALCON_TOPOX _TS1 |
CERT: 1.628E-5 |
0.5386 |
 |
 |
| Atome2_CBS_T S5 |
CERT: 1.692E-5 |
0.5377 |
 |
 |
| Seok-server_ TS4 |
CERT: 1.715E-5 |
0.5374 |
 |
 |
| HHPred1_TS1 | CERT: 1.728E-5 |
0.5372 |
 |
 |
| FALCON_TOPO_ TS2 |
CERT: 2.042E-5 |
0.5334 |
 |
 |
| slbio_TS2 | CERT: 2.086E-5 |
0.5329 |
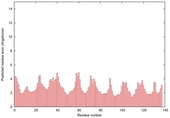 |
 |
| Seok-server_ TS2 |
CERT: 2.261E-5 |
0.5310 |
 |
 |
| IntFOLD4_TS1 |
CERT: 2.266E-5 |
0.5309 |
 |
 |
| QUARK_TS1 | CERT: 2.292E-5 |
0.5307 |
 |
 |
| Seok-server_ TS3 |
CERT: 2.386E-5 |
0.5297 |
 |
 |
| Seok-server_ TS1 |
CERT: 2.508E-5 |
0.5286 |
 |
 |
| myprotein-me _TS4 |
CERT: 2.704E-5 |
0.5269 |
 |
 |
| Seok-server_ TS5 |
CERT: 2.743E-5 |
0.5265 |
 |
 |
| YASARA_TS2 | CERT: 2.983E-5 |
0.5246 |
 |
 |
| Seok-assembl y_TS1 |
CERT: 3.081E-5 |
0.5238 |
 |
 |
| YASARA_TS1 | CERT: 3.163E-5 |
0.5232 |
 |
 |
| MUfold1_TS1 | CERT: 3.209E-5 |
0.5229 |
 |
 |
| ToyPred_emai l_TS1 |
CERT: 3.242E-5 |
0.5227 |
 |
 |
| MULTICOM-NOV EL_TS1 |
CERT: 3.493E-5 |
0.5209 |
 |
 |
| Seok-assembl y_TS4 |
CERT: 3.679E-5 |
0.5197 |
 |
 |
| Seok-assembl y_TS5 |
CERT: 3.69E-5 |
0.5197 |
 |
 |
| IntFOLD4_TS3 |
CERT: 3.691E-5 |
0.5197 |
 |
 |
| myprotein-me _TS2 |
CERT: 4.08E-5 |
0.5173 |
 |
 |
| IntFOLD4_TS2 |
CERT: 4.235E-5 |
0.5165 |
 |
 |
| Atome2_CBS_T S4 |
CERT: 4.366E-5 |
0.5158 |
 |
 |
| RaptorX_TS1 | CERT: 4.396E-5 |
0.5156 |
 |
 |
| myprotein-me _TS5 |
CERT: 4.403E-5 |
0.5156 |
 |
 |
| MULTICOM-NOV EL_TS2 |
CERT: 4.567E-5 |
0.5147 |
 |
 |
| MULTICOM-NOV EL_TS3 |
CERT: 4.796E-5 |
0.5136 |
 |
 |
| Seok-assembl y_TS3 |
CERT: 5.078E-5 |
0.5123 |
 |
 |
| RBO_Aleph_TS 2 |
CERT: 5.317E-5 |
0.5112 |
 |
 |
| MULTICOM-CON STRUCT_TS1 |
CERT: 5.395E-5 |
0.5109 |
 |
 |
| MUfold1_TS4 | CERT: 5.678E-5 |
0.5097 |
 |
 |
| myprotein-me _TS3 |
CERT: 5.743E-5 |
0.5094 |
 |
 |
| BhageerathH- Plus_TS1 |
CERT: 5.989E-5 |
0.5085 |
 |
 |
| MULTICOM-NOV EL_TS4 |
CERT: 6.104E-5 |
0.5080 |
 |
 |
| RBO_Aleph_TS 5 |
CERT: 6.176E-5 |
0.5078 |
 |
 |
| MULTICOM-CLU STER_TS5 |
CERT: 6.247E-5 |
0.5075 |
 |
 |
| MULTICOM-CLU STER_TS4 |
CERT: 6.355E-5 |
0.5071 |
 |
 |
| RBO_Aleph_TS 1 |
CERT: 6.445E-5 |
0.5068 |
 |
 |
| MULTICOM-CLU STER_TS3 |
CERT: 6.496E-5 |
0.5066 |
 |
 |
| RBO_Aleph_TS 4 |
CERT: 7.234E-5 |
0.5041 |
 |
 |
| myprotein-me _TS1 |
CERT: 7.465E-5 |
0.5034 |
 |
 |
| MUfold2_TS1 | CERT: 7.553E-5 |
0.5031 |
 |
 |
| MULTICOM-CLU STER_TS1 |
CERT: 7.576E-5 |
0.5030 |
 |
 |
| MUfold1_TS5 | CERT: 8.347E-5 |
0.5008 |
 |
 |
| RBO_Aleph_TS 3 |
CERT: 8.665E-5 |
0.4999 |
 |
 |
| MUfold1_TS3 | CERT: 9.138E-5 |
0.4987 |
 |
 |
| MULTICOM-CON STRUCT_TS2 |
CERT: 9.241E-5 |
0.4984 |
 |
 |
| MULTICOM-CON STRUCT_TS5 |
CERT: 9.531E-5 |
0.4977 |
 |
 |
| Seok-naive_a ssembly_TS1 |
CERT: 1.001E-4 |
0.4966 |
 |
 |
| MUfold1_TS2 | CERT: 1.11E-4 |
0.4942 |
 |
 |
| MUfold2_TS3 | CERT: 1.143E-4 |
0.4935 |
 |
 |
| MULTICOM-CON STRUCT_TS3 |
CERT: 1.197E-4 |
0.4924 |
 |
 |
| MULTICOM-CON STRUCT_TS4 |
CERT: 1.302E-4 |
0.4905 |
 |
 |
| MUfold2_TS2 | CERT: 1.398E-4 |
0.4889 |
 |
 |
| Seok-assembl y_TS2 |
CERT: 1.998E-4 |
0.4806 |
 |
 |
| MULTICOM-CLU STER_TS2 |
CERT: 2.212E-4 |
0.4782 |
 |
 |
| FLOUDAS_SERV ER_TS3 |
MEDIUM: 1.971E-2 |
0.3743 |
 |
 |
| FLOUDAS_SERV ER_TS1 |
MEDIUM: 2.194E-2 |
0.3720 |
 |
 |
| FLOUDAS_SERV ER_TS2 |
MEDIUM: 2.431E-2 |
0.3696 |
 |
 |
| FLOUDAS_SERV ER_TS5 |
MEDIUM: 2.708E-2 |
0.3670 |
 |
 |
| FLOUDAS_SERV ER_TS4 |
MEDIUM: 2.81E-2 |
0.3661 |
 |
 |
| Distill_TS5 | MEDIUM: 3.756E-2 |
0.3585 |
 |
 |
| Distill_TS1 | MEDIUM: 4.189E-2 |
0.3553 |
 |
 |
| Distill_TS2 | MEDIUM: 4.204E-2 |
0.3552 |
 |
 |
| Distill_TS4 | LOW: 6.692E-2 |
0.3396 |
 |
 |
| MULTICOM-NOV EL_TS5 |
LOW: 7.198E-2 |
0.3369 |
 |
 |
| Pcons-net_TS 3 |
POOR: 1.083E-1 |
0.3211 |
 |
 |
| Distill_TS3 | POOR: 1.128E-1 |
0.3195 |
 |
 |
| Pareto-serve r_TS1 |
POOR: 1.964E-1 |
0.2970 |
 |
 |
| chuo-u-serve r_TS4 |
POOR: 2.145E-1 |
0.2932 |
 |
 |
| chuo-u2_TS4 | POOR: 2.145E-1 |
0.2932 |
 |
 |
| Pcons-net_TS 1 |
POOR: 2.771E-1 |
0.2821 |
 |
 |
| FFAS-3D_TS1 | POOR: 3.567E-1 |
0.2701 |
 |
 |
| YASARA_TS5 | POOR: 3.583E-1 |
0.2699 |
 |
 |
| GAPF_LNCC_SE RVER_TS1 |
POOR: 3.603E-1 |
0.2696 |
 |
 |
| Pcons-net_TS 4 |
POOR: 3.698E-1 |
0.2683 |
 |
 |
| YASARA_TS4 | POOR: 3.984E-1 |
0.2644 |
 |
 |
| QUARK_TS4 | POOR: 3.995E-1 |
0.2642 |
 |
 |
| Zhang-Server _TS4 |
POOR: 4.027E-1 |
0.2638 |
 |
 |
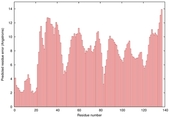
| Zhang-Server _TS5 |
POOR: 4.133E-1 |
0.2624 |
 |
 |
| PhyreTopoAlp ha_TS1 |
POOR: 4.215E-1 |
0.2613 |
 |
 |
| QUARK_TS3 | POOR: 4.316E-1 |
0.2600 |
 |
 |
| PhyreTopoAlp ha_TS2 |
POOR: 4.462E-1 |
0.2581 |
 |
 |
| PhyreTopoAlp ha_TS3 |
POOR: 4.701E-1 |
0.2551 |
 |
 |
| GAPF_LNCC_SE RVER_TS5 |
POOR: 5.039E-1 |
0.2508 |
 |
 |
| Pcons-net_TS 5 |
POOR: 5.105E-1 |
0.2500 |
 |
 |
| chuo-u-serve r_TS1 |
POOR: 5.495E-1 |
0.2451 |
 |
 |
| chuo-u2_TS1 | POOR: 5.495E-1 |
0.2451 |
 |
 |
| GAPF_LNCC_SE RVER_TS4 |
POOR: 5.668E-1 |
0.2429 |
 |
 |
| PhyreTopoAlp ha_TS5 |
POOR: 5.728E-1 |
0.2421 |
 |
 |
| Pareto-serve r_TS5 |
POOR: 5.784E-1 |
0.2414 |
 |
 |
| tsspred2_TS1 |
POOR: 5.976E-1 |
0.2390 |
 |
 |
| tsspred2_TS4 |
POOR: 6.005E-1 |
0.2386 |
 |
 |
| MULTICOM-REF INE_TS3 |
POOR: 6.223E-1 |
0.2358 |
 |
 |
| YASARA_TS3 | POOR: 6.388E-1 |
0.2336 |
 |
 |
| RaptorX-Cont act_TS1 |
POOR: 6.989E-1 |
0.2255 |
 |
 |
| tsspred2_TS3 |
POOR: 7.067E-1 |
0.2243 |
 |
 |
| RaptorX-Cont act_TS2 |
POOR: 7.104E-1 |
0.2238 |
 |
 |
| MULTICOM-REF INE_TS4 |
POOR: 7.146E-1 |
0.2232 |
 |
 |
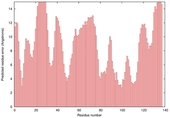
| RaptorX-Cont act_TS5 |
POOR: 7.654E-1 |
0.2155 |
 |
 |
| RaptorX-Cont act_TS3 |
POOR: 8.285E-1 |
0.2043 |
 |
 |
| RaptorX-Cont act_TS4 |
POOR: 8.293E-1 |
0.2041 |
 |
 |
| Seok-naive_a ssembly_TS4 |
POOR: 9.38E-1 |
0.1709 |
 |
 |
| Seok-naive_a ssembly_TS2 |
POOR: 9.404E-1 |
0.1695 |
 |
 |
| M4T-SmotifTF _TS4 |
POOR: 9.589E-1 |
0.1558 |
 |
 |
| M4T-SmotifTF _TS5 |
POOR: 9.62E-1 |
0.1527 |
 |
 |
| M4T-SmotifTF _TS2 |
POOR: 9.636E-1 |
0.1511 |
 |
 |
| M4T-SmotifTF _TS3 |
POOR: 9.649E-1 |
0.1497 |
 |
 |
| M4T-SmotifTF _TS1 |
POOR: 9.657E-1 |
0.1489 |
 |
 |