The ModFOLD Server Results
Please cite the following papers:Maghrabi, A.H.A. & McGuffin, L.J. (2017) ModFOLD6: an accurate web server for the global and local quality estimation of 3D models of proteins, 45, W416-W421, Nucleic Acids Res. doi: 10.1093/nar/gkx332. PubMed
McGuffin, L.J., Shuid, A.M., Kempster, R., Maghrabi, A.H.A., Nealon J.O., Salehe, B.R., Atkins, J.D. & Roche, D.B. (2017) Accurate Template Based Modelling in CASP12 using the IntFOLD4-TS, ModFOLD6 and ReFOLD methods. Proteins: Structure, Function, and Bioinformatics, 86 Suppl 1, 335-344, doi: 10.1002/prot.25360. PubMed
Click on the links below to download machine readable results files:
- Download Quality Assessment file (raw model quality prediction scores in CASP QA format)
- Make a compressed file containing all evaluated 3D models (models are in PDB format with predicted per-residue errors in the B-factor column)
Results will be available for 21 days after job completion (subject to server capacity)
| Graphical ModFOLD6 results for T0859 | Help | ||||
| Model name | Confidence and P-value | Global model quality score | Residue error plot (click image for large version) | 3D view of residue error (click image for large version) |
| server19_TS1 |
HIGH: 7.89E-3 |
0.3953 |
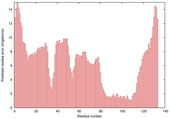 |
 |
| server07_TS1 |
LOW: 5.964E-2 |
0.3438 |
 |
 |
| server12_TS1 |
LOW: 8.314E-2 |
0.3315 |
 |
 |
| server06_TS1 |
POOR: 2.24E-1 |
0.2914 |
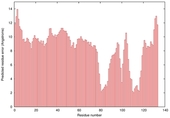 |
 |
| server15_TS1 |
POOR: 2.873E-1 |
0.2804 |
 |
 |
| server18_TS1 |
POOR: 3.333E-1 |
0.2734 |
 |
 |
| server11_TS1 |
POOR: 4.279E-1 |
0.2605 |
 |
 |
| server08_TS1 |
POOR: 4.718E-1 |
0.2548 |
 |
 |
| server17_TS1 |
POOR: 4.759E-1 |
0.2543 |
 |
 |
| server14_TS1 |
POOR: 5.068E-1 |
0.2504 |
 |
 |
| server09_TS1 |
POOR: 5.436E-1 |
0.2458 |
 |
 |
| server05_TS1 |
POOR: 5.998E-1 |
0.2387 |
 |
 |
| server01_TS1 |
POOR: 6.068E-1 |
0.2378 |
 |
 |
| server10_TS1 |
POOR: 6.169E-1 |
0.2365 |
 |
 |
| server16_TS1 |
POOR: 6.892E-1 |
0.2268 |
 |
 |
| server20_TS1 |
POOR: 7.214E-1 |
0.2222 |
 |
 |
| server04_TS1 |
POOR: 7.689E-1 |
0.2149 |
 |
 |
| server03_TS1 |
POOR: 7.893E-1 |
0.2115 |
 |
 |
| server02_TS1 |
POOR: 8.299E-1 |
0.2040 |
 |
 |
| server13_TS1 |
POOR: 8.594E-1 |
0.1976 |
 |
 |