The IntFOLD Server Results Page (Version 6.0)
Please cite the relevant papers:McGuffin, L.J., Adiyaman, R., Maghrabi, A.H.A., Shuid, A.N., Brackenridge, D.A., Nealon, J.O. & Philomina, L.S. (2019) IntFOLD: an integrated web resource for high performance protein structure and function prediction. Nucleic Acids Research, 47, W408-W413. DOI PubMed
McGuffin, L.J., Shuid, A.M., Kempster, R., Maghrabi, A.H.A., Nealon J.O., Salehe, B.R., Atkins, J.D. & Roche, D.B. (2017) Accurate Template Based Modelling in CASP12 using the IntFOLD4-TS, ModFOLD6 and ReFOLD methods. Proteins: Structure, Function, and Bioinformatics, 86 Suppl 1, 335-344, doi: 10.1002/prot.25360. PubMed
Links to graphical output:
- Top 5 3D models
- Disorder prediction
- Domain boundary prediction
- Binding site prediction
- Full model quality assessment results
Download machine readable results in CASP format:
- TS (Tertiary Structure Prediction)
- DR (Disorder Prediction)
- DP (Domain Prediction)
- FN (Binding Site Prediction)
- QA (Model Quality Prediction)
- Make a compressed file containing all generated 3D models (models are in PDB format with predicted per-residue errors in the B-factor column)
Results will be available for 21 days after job completion (subject to server capacity)
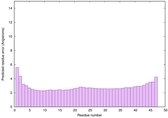
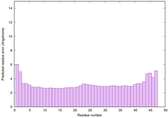
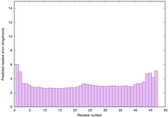
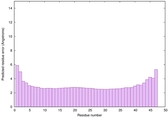
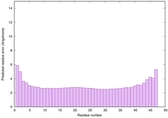
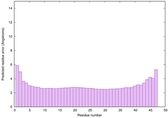
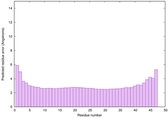
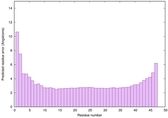
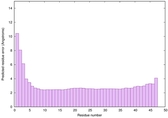
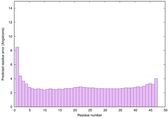
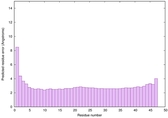
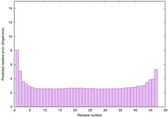
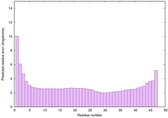
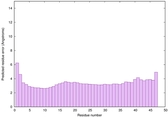
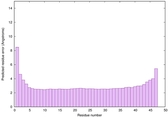
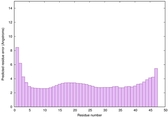
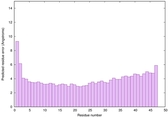
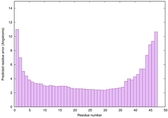
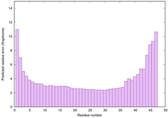
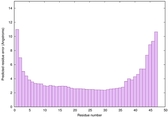
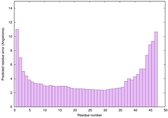
| Tertiary Structure Predictions: Top 5 multi-template 3D models for striatin-4 | Help | ||||
| Model ID and PDBsum links for all templates used | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | 3D map of model quality |
|
IntFOLD6 IntFOLDTS60 multi4 TS1 4n6jA 4rsiB |
CERT: 5.136E-17 |
0.6810 |
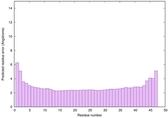 |

|
|
IntFOLD6 wMUSTER multi3 TS1 4n6jA 5vr2A 3he5A |
CERT: 8.443E-17 |
0.6788 |
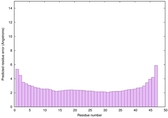 |

|
|
IntFOLD6 HHpred TS1 4n6jA |
CERT: 1.181E-16 |
0.6773 |
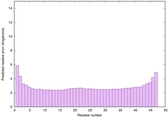 |

|
|
IntFOLD6 IT5 TS1 |
CERT: 1.247E-16 |
0.6771 |
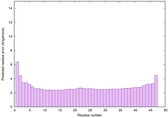 |

|
|
IntFOLD6 wMUSTER multi7 TS1 6h9mA2 3he5A |
CERT: 1.275E-16 |
0.6770 |
 |

|
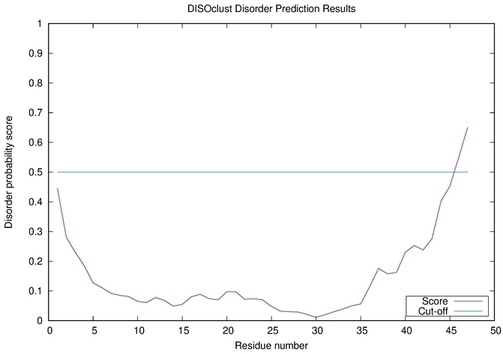
McGuffin, L. J. (2008) Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics, 24, 1789-1804. PubMed
| Disorder prediction for striatin-4 | Help | |||
| |||
| Click image to download plot in PostScript format. |
| Domain boundary prediction for striatin-4 | Help | |||

| |||
| Click image to download model or view prediction using Jmol. The model above is coloured according to the predicted domains. |
Roche, D. B., Tetchner, S. J. & McGuffin, L. J. (2011) FunFOLD: an improved automated method for the prediction of ligand binding residues using 3D models of proteins. BMC Bioinformatics, 12, 160. PubMed
| Ligand binding residues prediction for striatin-4 | Help | |||
| |||
| No binding residues predicted. |
Maghrabi, A.H.A. & McGuffin L.J. (2017) ModFOLD6: an accurate web server for the global and local quality estimation of 3D models of proteins. Nucleic Acids Res., 45, W416-W421, doi: 10.1093/nar/gkx332. PubMed
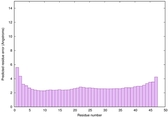
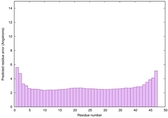
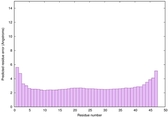
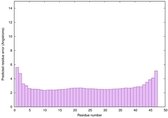
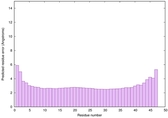
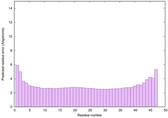
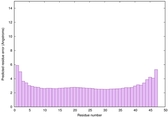
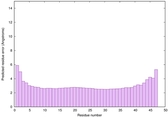
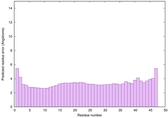
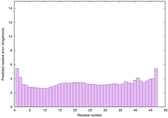
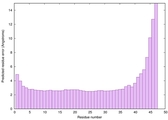
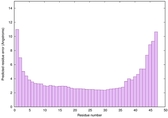
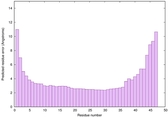
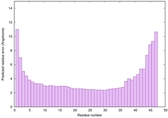
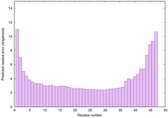
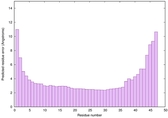
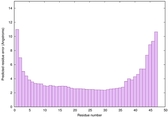
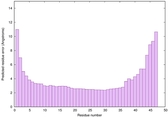
| Full model quality assessment results for striatin-4 | Help | ||||
| Model ID and PDBsum links for all templates used | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | 3D map of model quality |
|
IntFOLD6 IntFOLDTS60 multi4 TS1 4n6jA 4rsiB |
CERT: 5.136E-17 |
0.6810 |
 |

|
|
IntFOLD6 wMUSTER multi3 TS1 4n6jA 5vr2A 3he5A |
CERT: 8.443E-17 |
0.6788 |
 |

|
|
IntFOLD6 HHpred TS1 4n6jA |
CERT: 1.181E-16 |
0.6773 |
 |

|
|
IntFOLD6 IT5 TS1 |
CERT: 1.247E-16 |
0.6771 |
 |

|
|
IntFOLD6 wMUSTER multi7 TS1 6h9mA2 3he5A |
CERT: 1.275E-16 |
0.6770 |
 |

|
|
IntFOLD6 wMUSTER multi5 TS1 6h9mA2 3he5A |
CERT: 1.275E-16 |
0.6770 |
 |

|
|
IntFOLD6 PPAS multi5 TS1 6h9mA2 3he5A |
CERT: 1.275E-16 |
0.6770 |
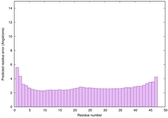 |

|
|
IntFOLD6 MUSTER multi7 TS1 6h9mA2 3he5A |
CERT: 1.275E-16 |
0.6770 |
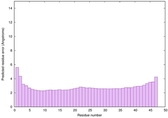 |

|
|
IntFOLD6 MUSTER multi5 TS1 6h9mA2 3he5A |
CERT: 1.275E-16 |
0.6770 |
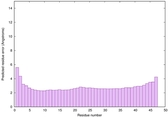 |

|
|
IntFOLD6 dPPAS multi5 TS1 6h9mA2 3he5A |
CERT: 1.275E-16 |
0.6770 |
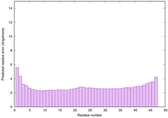 |

|
|
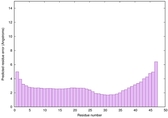
IntFOLD6 CNFsearch multi8 TS1 4e40A |
CERT: 1.296E-16 |
0.6769 |
 |
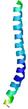
|
|
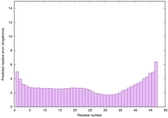
IntFOLD6 CNFsearch multi7 TS1 4e40A |
CERT: 1.296E-16 |
0.6769 |
 |
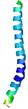
|
|
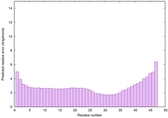
IntFOLD6 CNFsearch multi6 TS1 4e40A |
CERT: 1.296E-16 |
0.6769 |
 |
|
|
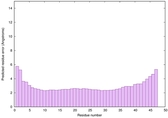
IntFOLD6 CNFsearch multi5 TS1 4e40A 4wheA |
CERT: 1.485E-16 |
0.6763 |
 |

|
|
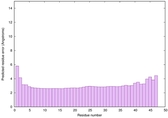
IntFOLD6 PPAS multi7 TS1 6h9mA2 3he5A 5al6A |
CERT: 3.343E-16 |
0.6727 |
 |
|
|
IntFOLD6 HHsearch multi8 TS1 4n6jA 4ywzA |
CERT: 3.448E-16 |
0.6726 |
 |

|
|
IntFOLD6 HHsearch multi5 TS1 4n6jA 4ywzA |
CERT: 3.448E-16 |
0.6726 |
 |

|
|
IntFOLD6 HHsearch multi4 TS1 4n6jA 4ywzA |
CERT: 3.448E-16 |
0.6726 |
 |

|
|
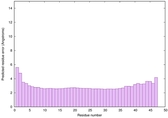
IntFOLD6 wPPAS multi7 TS1 3he5A 4pxjA |
CERT: 4.226E-16 |
0.6717 |
 |

|
|
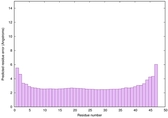
IntFOLD6 IntFOLDTS60 multi3 TS1 4n6jA 4ywzA 1yhnB 2do1A |
CERT: 4.718E-16 |
0.6712 |
 |

|
|
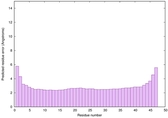
IntFOLD6 HHpred TS3 4n6jA |
CERT: 4.727E-16 |
0.6712 |
 |

|
|
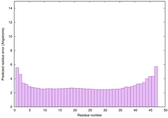
IntFOLD6 HHsearch multi7 TS1 4n6jA 4ywzA 1yhnB 2do1A |
CERT: 6.063E-16 |
0.6701 |
 |

|
|
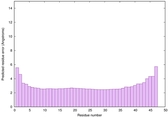
IntFOLD6 HHsearch multi3 TS1 4n6jA 4ywzA 1yhnB 2do1A |
CERT: 6.063E-16 |
0.6701 |
 |

|
|
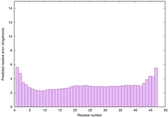
IntFOLD6 CNFsearch multi2 TS1 4n6jA |
CERT: 7.128E-16 |
0.6694 |
 |

|
|
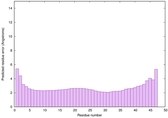
IntFOLD6 wPPAS multi4 TS1 4n6jA 5vr2A |
CERT: 7.22E-16 |
0.6693 |
 |

|
|
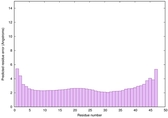
IntFOLD6 wPPAS multi1 TS1 4n6jA 5vr2A |
CERT: 7.22E-16 |
0.6693 |
 |

|
|
IntFOLD6 wMUSTER multi4 TS1 4n6jA 5vr2A |
CERT: 7.22E-16 |
0.6693 |
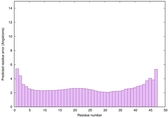 |

|
|
IntFOLD6 wMUSTER multi1 TS1 4n6jA 5vr2A |
CERT: 7.22E-16 |
0.6693 |
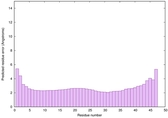 |

|
|
IntFOLD6 CNFsearch multi4 TS1 4n6jA 4wheA |
CERT: 1.01E-15 |
0.6678 |
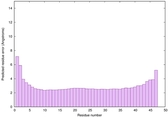 |

|
|
IntFOLD6 CNFsearch multi1 TS1 4n6jA 4wheA |
CERT: 1.01E-15 |
0.6678 |
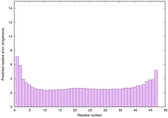 |

|
|
IntFOLD6 sp3 multi5 TS1 3he5B 4n6jA |
CERT: 1.287E-15 |
0.6668 |
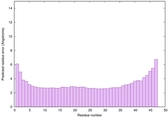 |

|
|
IntFOLD6 CNFsearch multi3 TS1 4n6jA 4wheA 4e40A |
CERT: 1.346E-15 |
0.6666 |
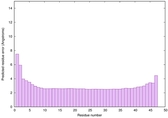 |

|
|
IntFOLD6 sp3 multi1 TS1 4n6jA 2oqqA |
CERT: 2.404E-15 |
0.6640 |
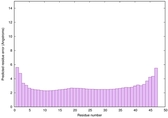 |

|
|
IntFOLD6 SPARKSX multi7 TS1 4n6jA |
CERT: 2.723E-15 |
0.6635 |
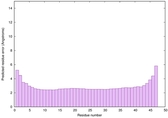 |

|
|
IntFOLD6 SPARKSX multi6 TS1 4n6jA |
CERT: 2.723E-15 |
0.6635 |
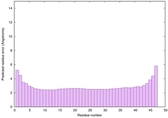 |

|
|
IntFOLD6 SPARKSX multi3 TS1 4n6jA |
CERT: 2.723E-15 |
0.6635 |
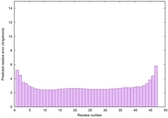 |

|
|
IntFOLD6 SPARKSX multi2 TS1 4n6jA |
CERT: 2.723E-15 |
0.6635 |
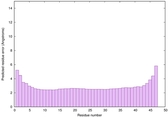 |

|
|
IntFOLD6 wPPAS multi3 TS1 4n6jA 5vr2A 2oqqA 4pxjA 3he5A |
CERT: 2.774E-15 |
0.6634 |
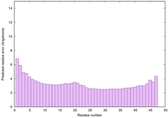 |

|
|
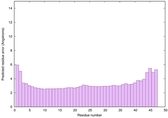
IntFOLD6 PPAS multi3 TS1 4n6jA 5vr2A 2oqqA 4pkyB 3he5A 5al6A |
CERT: 2.963E-15 |
0.6631 |
 |

|
|
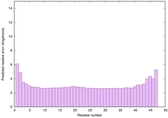
IntFOLD6 dPPAS multi7 TS1 6h9mA2 3he5A 5al6A 5kb0A |
CERT: 3.047E-15 |
0.6630 |
 |

|
|
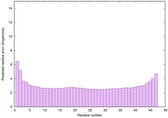
IntFOLD6 wdPPAS multi3 TS1 4n6jA 5vr2A 2oqqA 4pxjA 3he5A 5kb0A 5al6A |
CERT: 3.579E-15 |
0.6623 |
 |

|
|
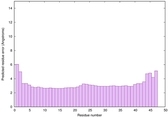
IntFOLD6 spk2 multi8 TS1 5vr2A |
CERT: 3.922E-15 |
0.6619 |
 |

|
|
IntFOLD6 spk2 multi7 TS1 5vr2A |
CERT: 3.922E-15 |
0.6619 |
 |

|
|
IntFOLD6 spk2 multi6 TS1 5vr2A |
CERT: 3.922E-15 |
0.6619 |
 |

|
|
IntFOLD6 IntFOLDTS60 multi2 TS1 4n6jA |
CERT: 4.061E-15 |
0.6617 |
 |

|
|
IntFOLD6 IntFOLDTS60 multi1 TS1 4n6jA |
CERT: 4.061E-15 |
0.6617 |
 |

|
|
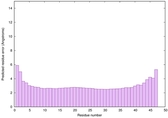
IntFOLD6 HHsearch multi6 TS1 4n6jA |
CERT: 4.061E-15 |
0.6617 |
 |

|
|
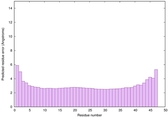
IntFOLD6 HHsearch multi2 TS1 4n6jA |
CERT: 4.061E-15 |
0.6617 |
 |

|
|
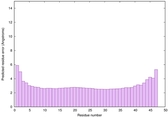
IntFOLD6 COMA multi8 TS1 4n6jA |
CERT: 4.061E-15 |
0.6617 |
 |

|
|
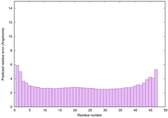
IntFOLD6 COMA multi7 TS1 4n6jA |
CERT: 4.061E-15 |
0.6617 |
 |

|
|
IntFOLD6 COMA multi6 TS1 4n6jA |
CERT: 4.061E-15 |
0.6617 |
 |

|
|
IntFOLD6 COMA multi5 TS1 4n6jA |
CERT: 4.061E-15 |
0.6617 |
 |

|
|
IntFOLD6 COMA multi4 TS1 4n6jA |
CERT: 4.061E-15 |
0.6617 |
 |

|
|
IntFOLD6 COMA multi3 TS1 4n6jA |
CERT: 4.061E-15 |
0.6617 |
 |

|
|
IntFOLD6 COMA multi2 TS1 4n6jA |
CERT: 4.061E-15 |
0.6617 |
 |

|
|
IntFOLD6 COMA multi1 TS1 4n6jA |
CERT: 4.061E-15 |
0.6617 |
 |

|
|
IntFOLD6 dPPAS2 multi7 TS1 4pkyB 3he5A 4pxjA 5al6A 5kb0A |
CERT: 5.423E-15 |
0.6604 |
 |

|
|
IntFOLD6 IntFOLDTS140 multi3 TS1 6h9mA2 3he5A 4pxjA 5al6A 5kb0A 4ywzA 1yhnB 2do1A |
CERT: 6.281E-15 |
0.6598 |
 |

|
|
IntFOLD6 wPPAS multi6 TS1 3he5A |
CERT: 8.134E-15 |
0.6587 |
 |

|
|
IntFOLD6 wdPPAS multi6 TS1 3he5A |
CERT: 8.134E-15 |
0.6587 |
 |

|
|
IntFOLD6 dPPAS2 multi3 TS1 4n6jA 5vr2A 4pxjA 5al6A |
CERT: 1.058E-14 |
0.6575 |
 |

|
|
IntFOLD6 sp3 multi4 TS1 4n6jA 3he5B |
CERT: 1.245E-14 |
0.6568 |
 |

|
|
IntFOLD6 dPPAS multi3 TS1 4n6jA 5vr2A 2oqqA 3he5A 5kb0A 5al6A |
CERT: 1.322E-14 |
0.6565 |
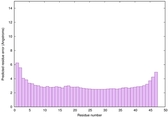 |

|
|
IntFOLD6 HHsearch multi1 TS1 4n6jA 4zqaA |
CERT: 1.335E-14 |
0.6565 |
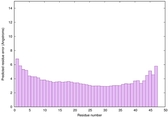 |

|
|
IntFOLD6 HHpred TS2 4n6jA |
CERT: 2.118E-14 |
0.6544 |
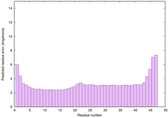 |

|
|
IntFOLD6 SPARKSX multi1 TS1 4n6jA 4hb1A |
CERT: 2.192E-14 |
0.6543 |
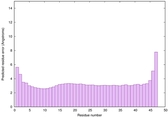 |

|
|
IntFOLD6 SPARKSX multi8 TS1 4n6jA 5vnyA |
CERT: 3.861E-14 |
0.6518 |
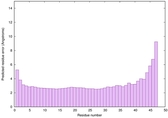 |

|
|
IntFOLD6 SPARKSX multi5 TS1 4n6jA 5vnyA |
CERT: 3.861E-14 |
0.6518 |
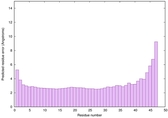 |

|
|
IntFOLD6 sp3 multi8 TS1 3he5B |
CERT: 6.485E-14 |
0.6495 |
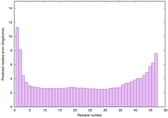 |

|
|
IntFOLD6 sp3 multi7 TS1 3he5B |
CERT: 6.485E-14 |
0.6495 |
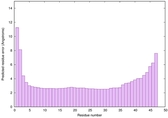 |

|
|
IntFOLD6 sp3 multi6 TS1 3he5B |
CERT: 6.485E-14 |
0.6495 |
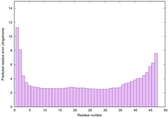 |

|
|
IntFOLD6 wdPPAS multi7 TS1 3he5A 4pxjA 5al6A 5kb0A |
CERT: 1.013E-13 |
0.6476 |
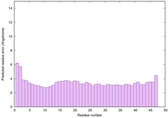 |

|
|
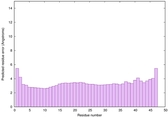
IntFOLD6 wMUSTER multi8 TS1 6h9mA2 |
CERT: 1.05E-13 |
0.6474 |
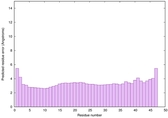 |
|
|
IntFOLD6 wMUSTER multi6 TS1 6h9mA2 |
CERT: 1.05E-13 |
0.6474 |
 |
|
|
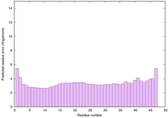
IntFOLD6 PPAS multi8 TS1 6h9mA2 |
CERT: 1.05E-13 |
0.6474 |
 |
|
|
IntFOLD6 PPAS multi6 TS1 6h9mA2 |
CERT: 1.05E-13 |
0.6474 |
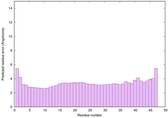 |
|
|
IntFOLD6 MUSTER multi8 TS1 6h9mA2 |
CERT: 1.05E-13 |
0.6474 |
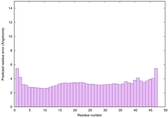 |
|
|
IntFOLD6 MUSTER multi6 TS1 6h9mA2 |
CERT: 1.05E-13 |
0.6474 |
 |
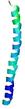
|
|
IntFOLD6 IntFOLDTS140 multi4 TS1 6h9mA2 |
CERT: 1.05E-13 |
0.6474 |
 |
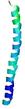
|
|
IntFOLD6 IntFOLDTS140 multi2 TS1 6h9mA2 |
CERT: 1.05E-13 |
0.6474 |
 |
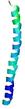
|
|
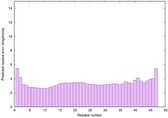
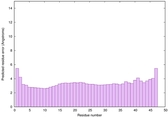
IntFOLD6 IntFOLDTS140 multi1 TS1 6h9mA2 |
CERT: 1.05E-13 |
0.6474 |
 |
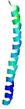
|
|
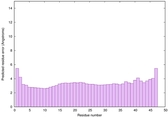
IntFOLD6 dPPAS multi8 TS1 6h9mA2 |
CERT: 1.05E-13 |
0.6474 |
 |
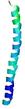
|
|
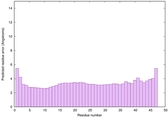
IntFOLD6 dPPAS multi6 TS1 6h9mA2 |
CERT: 1.05E-13 |
0.6474 |
 |
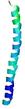
|
|
IntFOLD6 MUSTER multi3 TS1 4n6jA 5vr2A 3he5A |
CERT: 1.135E-13 |
0.6470 |
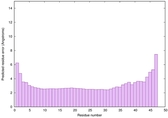 |

|
|
IntFOLD6 wPPAS multi8 TS1 3he5A 5vr2A |
CERT: 1.401E-13 |
0.6461 |
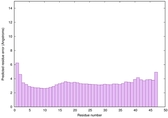 |

|
|
IntFOLD6 wPPAS multi5 TS1 3he5A 5vr2A |
CERT: 1.401E-13 |
0.6461 |
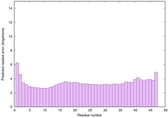 |

|
|
IntFOLD6 wdPPAS multi8 TS1 3he5A 5vr2A |
CERT: 1.401E-13 |
0.6461 |
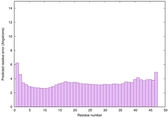 |

|
|
IntFOLD6 wdPPAS multi5 TS1 3he5A 5vr2A |
CERT: 1.401E-13 |
0.6461 |
 |

|
|
IntFOLD6 LOMETS multi3 TS1 4n6jA 5vr2A |
CERT: 1.704E-13 |
0.6453 |
 |

|
|
IntFOLD6 sp3 multi3 TS1 4n6jA 2oqqA 3he5B |
CERT: 3.426E-13 |
0.6422 |
 |

|
|
IntFOLD6 dPPAS2 multi8 TS1 4pkyB 5f5sB |
CERT: 2.089E-12 |
0.6342 |
 |

|
|
IntFOLD6 dPPAS2 multi5 TS1 4pkyB 5f5sB |
CERT: 2.089E-12 |
0.6342 |
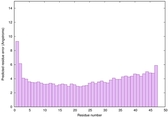 |

|
|
IntFOLD6 spk2 multi5 TS1 5vr2A 3he5B |
CERT: 2.625E-12 |
0.6332 |
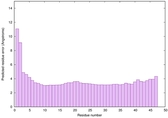 |

|
|
IntFOLD6 spk2 multi3 TS1 4n6jA 3he5B 5vr2A |
CERT: 2.932E-12 |
0.6327 |
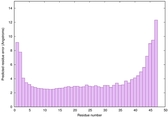 |

|
|
IntFOLD6 wdPPAS multi4 TS1 4n6jA 5vr2A |
CERT: 8.907E-12 |
0.6278 |
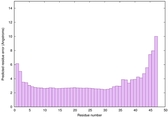 |

|
|
IntFOLD6 wdPPAS multi1 TS1 4n6jA 5vr2A |
CERT: 8.907E-12 |
0.6278 |
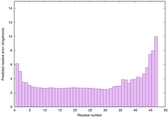 |

|
|
IntFOLD6 PPAS multi4 TS1 4n6jA 5vr2A |
CERT: 8.907E-12 |
0.6278 |
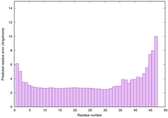 |

|
|
IntFOLD6 PPAS multi1 TS1 4n6jA 5vr2A |
CERT: 8.907E-12 |
0.6278 |
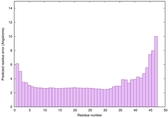 |

|
|
IntFOLD6 MUSTER multi4 TS1 4n6jA 5vr2A |
CERT: 8.907E-12 |
0.6278 |
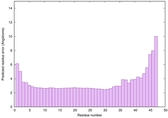 |

|
|
IntFOLD6 MUSTER multi1 TS1 4n6jA 5vr2A |
CERT: 8.907E-12 |
0.6278 |
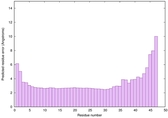 |

|
|
IntFOLD6 LOMETS multi4 TS1 4n6jA 5vr2A |
CERT: 8.907E-12 |
0.6278 |
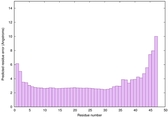 |

|
|
IntFOLD6 dPPAS multi4 TS1 4n6jA 5vr2A |
CERT: 8.907E-12 |
0.6278 |
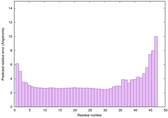 |

|
|
IntFOLD6 dPPAS multi1 TS1 4n6jA 5vr2A |
CERT: 8.907E-12 |
0.6278 |
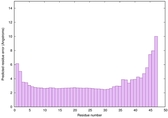 |

|
|
IntFOLD6 dPPAS2 multi4 TS1 4n6jA 5vr2A |
CERT: 8.907E-12 |
0.6278 |
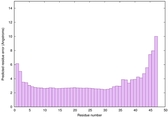 |

|
|
IntFOLD6 dPPAS2 multi1 TS1 4n6jA 5vr2A |
CERT: 8.907E-12 |
0.6278 |
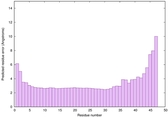 |

|
|
IntFOLD6 dPPAS2 multi6 TS1 4pkyB |
CERT: 6.753E-11 |
0.6189 |
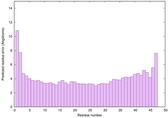 |

|
|
IntFOLD6 sp3 multi2 TS1 4n6jA |
CERT: 1.157E-10 |
0.6165 |
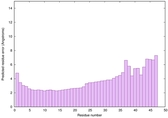 |

|
|
IntFOLD6 spk2 multi1 TS1 4n6jA 2oqqA |
CERT: 1.402E-10 |
0.6157 |
 |

|
|
IntFOLD6 wdPPAS multi2 TS1 4n6jA |
CERT: 1.43E-10 |
0.6156 |
 |

|
|
IntFOLD6 PPAS multi2 TS1 4n6jA |
CERT: 1.43E-10 |
0.6156 |
 |

|
|
IntFOLD6 MUSTER multi2 TS1 4n6jA |
CERT: 1.43E-10 |
0.6156 |
 |

|
|
IntFOLD6 LOMETS multi2 TS1 4n6jA |
CERT: 1.43E-10 |
0.6156 |
 |

|
|
IntFOLD6 LOMETS multi1 TS1 4n6jA |
CERT: 1.43E-10 |
0.6156 |
 |

|
|
IntFOLD6 Env-PPAS multi7 TS1 4n6jA |
CERT: 1.43E-10 |
0.6156 |
 |

|
|
IntFOLD6 Env-PPAS multi6 TS1 4n6jA |
CERT: 1.43E-10 |
0.6156 |
 |

|
|
IntFOLD6 Env-PPAS multi3 TS1 4n6jA |
CERT: 1.43E-10 |
0.6156 |
 |

|
|
IntFOLD6 Env-PPAS multi2 TS1 4n6jA |
CERT: 1.43E-10 |
0.6156 |
 |

|
|
IntFOLD6 dPPAS multi2 TS1 4n6jA |
CERT: 1.43E-10 |
0.6156 |
 |

|
|
IntFOLD6 dPPAS2 multi2 TS1 4n6jA |
CERT: 1.43E-10 |
0.6156 |
 |

|
|
IntFOLD6 wPPAS multi2 TS1 4n6jA |
CERT: 2.99E-10 |
0.6124 |
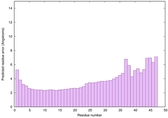 |

|
|
IntFOLD6 wMUSTER multi2 TS1 4n6jA |
CERT: 2.99E-10 |
0.6124 |
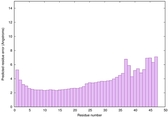 |

|
|
IntFOLD6 spk2 multi4 TS1 4n6jA 3he5B |
CERT: 2.295E-9 |
0.6034 |
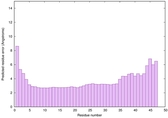 |

|
|
IntFOLD6 SPARKSX multi4 TS1 4n6jA 1mftB |
CERT: 4.395E-7 |
0.5802 |
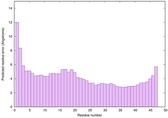 |

|
|
IntFOLD6 spk2 multi2 TS1 4n6jA |
CERT: 7.781E-7 |
0.5777 |
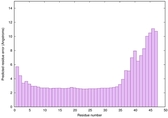 |

|
|
IntFOLD6 Env-PPAS multi8 TS1 4n6jA 3bbnO |
CERT: 9.074E-4 |
0.5466 |
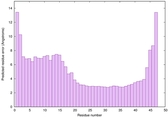 |
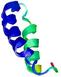
|
|
IntFOLD6 Env-PPAS multi5 TS1 4n6jA 3bbnO |
CERT: 9.074E-4 |
0.5466 |
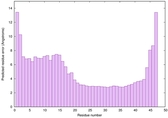 |
|
|
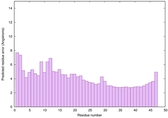
IntFOLD6 DMPf TS1 |
HIGH: 4.611E-3 |
0.5334 |
 |

|
|
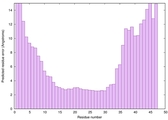
IntFOLD6 Env-PPAS multi4 TS1 4n6jA 5oeuA2 |
MEDIUM: 1.312E-2 |
0.5202 |
 |

|
|
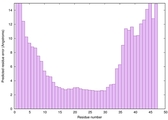
IntFOLD6 Env-PPAS multi1 TS1 4n6jA 5oeuA2 |
MEDIUM: 1.312E-2 |
0.5202 |
 |

|