The IntFOLD Server Results Page (Version 5.0)
Please cite the relevant papers:McGuffin, L.J., Adiyaman, R., Maghrabi, A.H.A., Shuid, A.N., Brackenridge, D.A., Nealon, J.O. & Philomina, L.S. (2019) IntFOLD: an integrated web resource for high performance protein structure and function prediction. Nucleic Acids Research, 47, W408-W413. DOI PubMed
McGuffin, L.J., Shuid, A.M., Kempster, R., Maghrabi, A.H.A., Nealon J.O., Salehe, B.R., Atkins, J.D. & Roche, D.B. (2017) Accurate Template Based Modelling in CASP12 using the IntFOLD4-TS, ModFOLD6 and ReFOLD methods. Proteins: Structure, Function, and Bioinformatics, 86 Suppl 1, 335-344, doi: 10.1002/prot.25360. PubMed
Links to graphical output:
- Top 5 3D models
- Disorder prediction
- Domain boundary prediction
- Binding site prediction
- Full model quality assessment results
Download machine readable results in CASP format:
- TS (Tertiary Structure Prediction)
- DR (Disorder Prediction)
- DP (Domain Prediction)
- FN (Binding Site Prediction)
- QA (Model Quality Prediction)
- Make a compressed file containing all generated 3D models (models are in PDB format with predicted per-residue errors in the B-factor column)
Results will be available for 21 days after job completion (subject to server capacity)
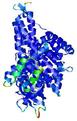
| Tertiary Structure Predictions: Top 5 multi-template 3D models for T0961 | Help | ||||
| Model ID and PDBsum links for all templates used | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | 3D map of model quality |
|
IntFOLD5 IntFOLDTS140 multi4 TS1 5ez3A 4y9jA |
CERT: 1.014E-9 |
0.6051 |
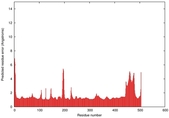 |
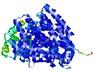
|
|
IntFOLD5 IntFOLDTS140 multi3 TS1 5ez3A 4y9jA |
CERT: 1.014E-9 |
0.6051 |
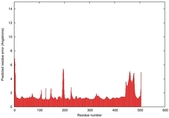 |
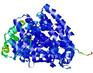
|
|
IntFOLD5 IntFOLDTS140 multi1 TS1 5ez3A 4y9jA |
CERT: 1.014E-9 |
0.6051 |
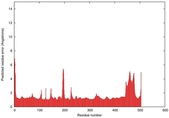 |
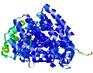
|
|
IntFOLD5 IntFOLDTS60 multi3 TS1 4y9jA 1bucA |
CERT: 1.015E-9 |
0.6050 |
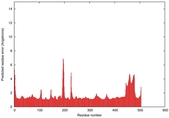 |
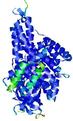
|
|
IntFOLD5 HHsearch multi1 TS1 4y9jA 3djlA |
CERT: 1.044E-9 |
0.6045 |
 |
|
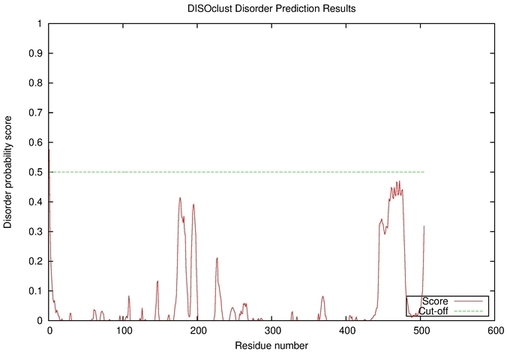
McGuffin, L. J. (2008) Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics, 24, 1789-1804. PubMed
| Disorder prediction for T0961 | Help | |||
| |||
| Click image to download plot in PostScript format. |
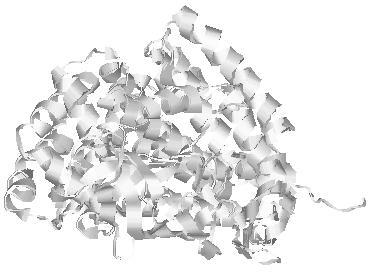
| Domain boundary prediction for T0961 | Help | |||
| |||
| Click image to download model or view prediction using Jmol. The model above is coloured according to the predicted domains. |
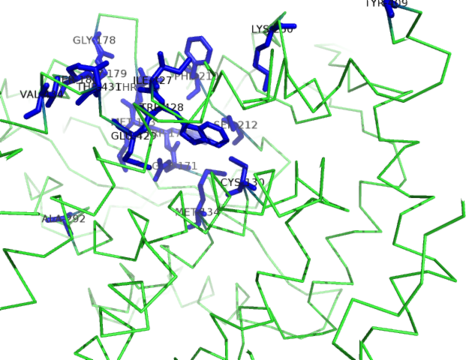
Roche, D. B., Tetchner, S. J. & McGuffin, L. J. (2011) FunFOLD: an improved automated method for the prediction of ligand binding residues using 3D models of proteins. BMC Bioinformatics, 12, 160. PubMed
| Ligand binding residues prediction for T0961 | Help | |||
| |||
| Click image to download model or view prediction using Jmol. Predicted ligand binding residues are shown as blue sticks in the image above. |
Maghrabi, A.H.A. & McGuffin L.J. (2017) ModFOLD6: an accurate web server for the global and local quality estimation of 3D models of proteins. Nucleic Acids Res., 45, W416-W421, doi: 10.1093/nar/gkx332. PubMed
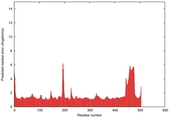
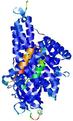
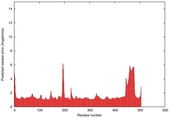
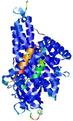
| Full model quality assessment results for T0961 | Help | ||||
| Model ID and PDBsum links for all templates used | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | 3D map of model quality |
|
IntFOLD5 IntFOLDTS140 multi4 TS1 5ez3A 4y9jA |
CERT: 1.014E-9 |
0.6051 |
 |

|
|
IntFOLD5 IntFOLDTS140 multi3 TS1 5ez3A 4y9jA |
CERT: 1.014E-9 |
0.6051 |
 |

|
|
IntFOLD5 IntFOLDTS140 multi1 TS1 5ez3A 4y9jA |
CERT: 1.014E-9 |
0.6051 |
 |

|
|
IntFOLD5 IntFOLDTS60 multi3 TS1 4y9jA 1bucA |
CERT: 1.015E-9 |
0.6050 |
 |

|
|
IntFOLD5 HHsearch multi1 TS1 4y9jA 3djlA |
CERT: 1.044E-9 |
0.6045 |
 |

|
|
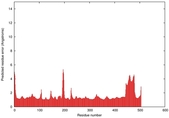
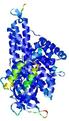
IntFOLD5 IntFOLDTS60 multi4 TS1 4y9jA |
CERT: 1.052E-9 |
0.6043 |
 |
|
|
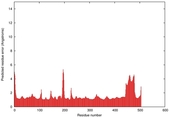
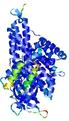
IntFOLD5 IntFOLDTS60 multi2 TS1 4y9jA |
CERT: 1.052E-9 |
0.6043 |
 |
|
|
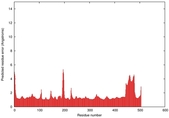
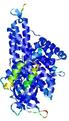
IntFOLD5 CNFsearch multi8 TS1 4y9jA |
CERT: 1.052E-9 |
0.6043 |
 |
|
|
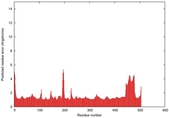
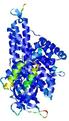
IntFOLD5 CNFsearch multi7 TS1 4y9jA |
CERT: 1.052E-9 |
0.6043 |
 |
|
|
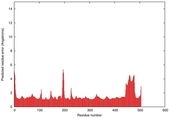
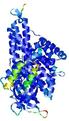
IntFOLD5 CNFsearch multi6 TS1 4y9jA |
CERT: 1.052E-9 |
0.6043 |
 |
|
|
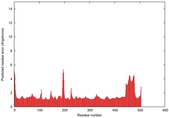
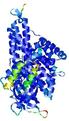
IntFOLD5 CNFsearch multi4 TS1 4y9jA |
CERT: 1.052E-9 |
0.6043 |
 |
|
|
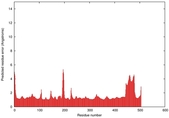
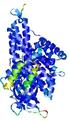
IntFOLD5 CNFsearch multi3 TS1 4y9jA |
CERT: 1.052E-9 |
0.6043 |
 |
|
|
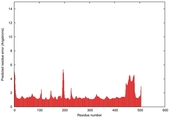
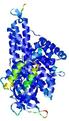
IntFOLD5 CNFsearch multi2 TS1 4y9jA |
CERT: 1.052E-9 |
0.6043 |
 |
|
|
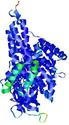
IntFOLD5 CNFsearch multi5 TS1 4y9jA 5ez3A |
CERT: 1.11E-9 |
0.6033 |
 |
|
|
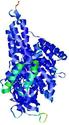
IntFOLD5 CNFsearch multi1 TS1 4y9jA 5ez3A |
CERT: 1.11E-9 |
0.6033 |
 |
|
|
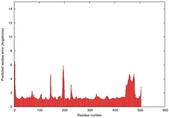
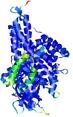
IntFOLD5 spk2 multi5 TS1 4y9jA 3djlA |
CERT: 1.139E-9 |
0.6028 |
 |
|
|
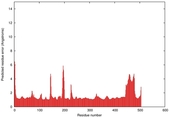
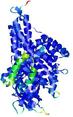
IntFOLD5 spk2 multi1 TS1 4y9jA 3djlA |
CERT: 1.139E-9 |
0.6028 |
 |
|
|
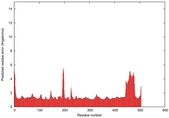
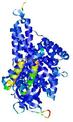
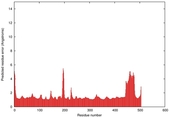
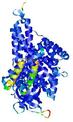
IntFOLD5 dPPAS2 multi7 TS1 4y9jA 4y9jA1 5ez3A2 |
CERT: 1.175E-9 |
0.6022 |
 |
|
|
IntFOLD5 dPPAS2 multi3 TS1 4y9jA 4y9jA1 5ez3A2 |
CERT: 1.175E-9 |
0.6022 |
 |
|
|
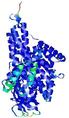
IntFOLD5 MUSTER multi5 TS1 4y9jA 5ez3A |
CERT: 1.188E-9 |
0.6019 |
 |
|
|
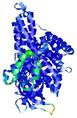
IntFOLD5 sp3 multi5 TS1 4y9jA 3djlA |
CERT: 1.23E-9 |
0.6013 |
 |
|
|
IntFOLD5 sp3 multi1 TS1 4y9jA 3djlA |
CERT: 1.23E-9 |
0.6013 |
 |
|
|
IntFOLD5 wMUSTER multi5 TS1 4y9jA 5ez3A |
CERT: 1.255E-9 |
0.6009 |
 |
|
|
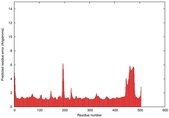
IntFOLD5 MUSTER multi8 TS1 4y9jA |
CERT: 1.258E-9 |
0.6008 |
 |
|
|
IntFOLD5 MUSTER multi6 TS1 4y9jA |
CERT: 1.258E-9 |
0.6008 |
 |
|
|
IntFOLD5 MUSTER multi4 TS1 4y9jA |
CERT: 1.258E-9 |
0.6008 |
 |
|
|
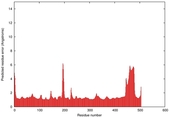
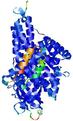
IntFOLD5 MUSTER multi2 TS1 4y9jA |
CERT: 1.258E-9 |
0.6008 |
 |
|
|
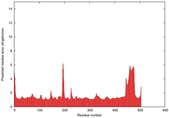
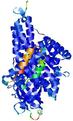
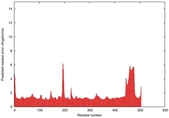
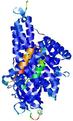
IntFOLD5 LOMETS multi4 TS1 4y9jA |
CERT: 1.258E-9 |
0.6008 |
 |
|
|
IntFOLD5 LOMETS multi2 TS1 4y9jA |
CERT: 1.258E-9 |
0.6008 |
 |
|
|
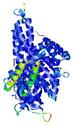
IntFOLD5 dPPAS multi3 TS1 4y9jA 4y9jA1 5ez3A2 |
CERT: 1.301E-9 |
0.6002 |
 |
|
|
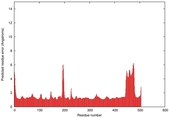
IntFOLD5 HHsearch multi8 TS1 4y9jA |
CERT: 1.304E-9 |
0.6001 |
 |

|
|
IntFOLD5 HHsearch multi7 TS1 4y9jA |
CERT: 1.304E-9 |
0.6001 |
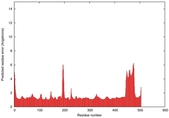 |

|
|
IntFOLD5 HHsearch multi6 TS1 4y9jA |
CERT: 1.304E-9 |
0.6001 |
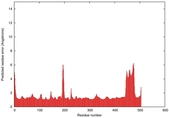 |

|
|
IntFOLD5 HHsearch multi4 TS1 4y9jA |
CERT: 1.304E-9 |
0.6001 |
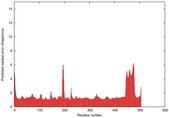 |

|
|
IntFOLD5 HHsearch multi3 TS1 4y9jA |
CERT: 1.304E-9 |
0.6001 |
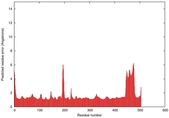 |

|
|
IntFOLD5 HHsearch multi2 TS1 4y9jA |
CERT: 1.304E-9 |
0.6001 |
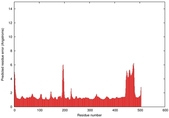 |

|
|
IntFOLD5 wdPPAS multi7 TS1 4y9jA 4y9jA1 5ez3A2 |
CERT: 1.33E-9 |
0.5997 |
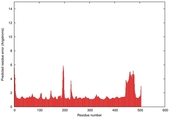 |
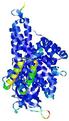
|
|
IntFOLD5 wdPPAS multi3 TS1 4y9jA 4y9jA1 5ez3A2 |
CERT: 1.33E-9 |
0.5997 |
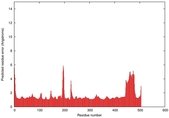 |
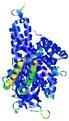
|
|
IntFOLD5 wMUSTER multi8 TS1 4y9jA |
CERT: 1.369E-9 |
0.5992 |
 |
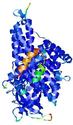
|
|
IntFOLD5 wMUSTER multi6 TS1 4y9jA |
CERT: 1.369E-9 |
0.5992 |
 |
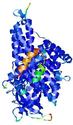
|
|
IntFOLD5 wMUSTER multi4 TS1 4y9jA |
CERT: 1.369E-9 |
0.5992 |
 |
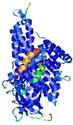
|
|
IntFOLD5 wMUSTER multi2 TS1 4y9jA |
CERT: 1.369E-9 |
0.5992 |
 |
|
|
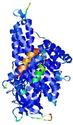
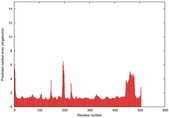
IntFOLD5 dPPAS2 multi5 TS1 4y9jA 5ez3A |
CERT: 1.378E-9 |
0.5991 |
 |
|
|
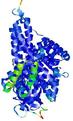
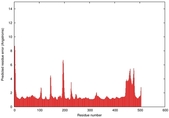
IntFOLD5 HHpred TS3 3djlA 4y9jA 5ez3A |
CERT: 1.384E-9 |
0.5990 |
 |

|
|
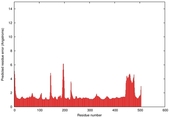
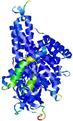
IntFOLD5 spk2 multi8 TS1 4y9jA |
CERT: 1.408E-9 |
0.5986 |
 |
|
|
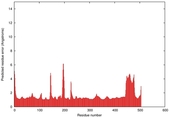
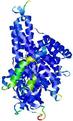
IntFOLD5 spk2 multi7 TS1 4y9jA |
CERT: 1.408E-9 |
0.5986 |
 |
|
|
IntFOLD5 spk2 multi6 TS1 4y9jA |
CERT: 1.408E-9 |
0.5986 |
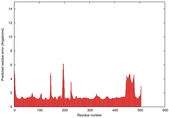 |
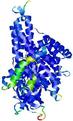
|
|
IntFOLD5 spk2 multi4 TS1 4y9jA |
CERT: 1.408E-9 |
0.5986 |
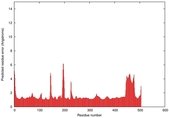 |
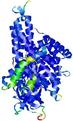
|
|
IntFOLD5 spk2 multi3 TS1 4y9jA |
CERT: 1.408E-9 |
0.5986 |
 |
|
|
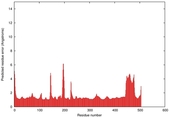
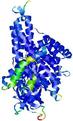
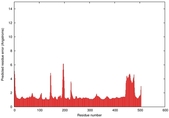
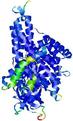
IntFOLD5 spk2 multi2 TS1 4y9jA |
CERT: 1.408E-9 |
0.5986 |
 |
|
|
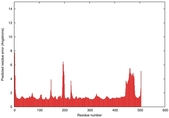
IntFOLD5 dPPAS multi7 TS1 5ez3A 4y9jA 4y9jA1 5ez3A2 |
CERT: 1.412E-9 |
0.5986 |
 |

|
|
IntFOLD5 wMUSTER multi7 TS1 4y9jA 4y9jA1 |
CERT: 1.416E-9 |
0.5985 |
 |
|
|
IntFOLD5 wMUSTER multi3 TS1 4y9jA 4y9jA1 |
CERT: 1.416E-9 |
0.5985 |
 |
|
|
IntFOLD5 wMUSTER multi1 TS1 4y9jA 4y9jA1 |
CERT: 1.416E-9 |
0.5985 |
 |
|
|
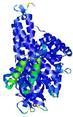
IntFOLD5 Env-PPAS multi5 TS1 4y9jA 5ez3A |
CERT: 1.469E-9 |
0.5978 |
 |
|
|
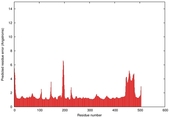
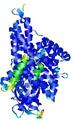
IntFOLD5 IntFOLDTS60 multi1 TS1 4y9jA |
CERT: 1.47E-9 |
0.5978 |
 |
|
|
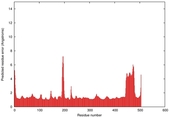
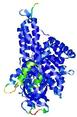
IntFOLD5 COMA multi8 TS1 4y9jA |
CERT: 1.485E-9 |
0.5976 |
 |
|
|
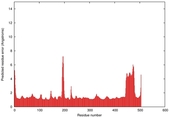
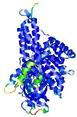
IntFOLD5 COMA multi7 TS1 4y9jA |
CERT: 1.485E-9 |
0.5976 |
 |
|
|
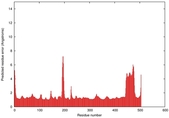
IntFOLD5 COMA multi6 TS1 4y9jA |
CERT: 1.485E-9 |
0.5976 |
 |
|
|
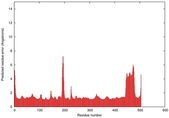
IntFOLD5 COMA multi4 TS1 4y9jA |
CERT: 1.485E-9 |
0.5976 |
 |
|
|
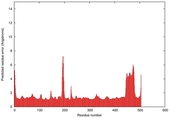
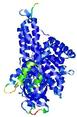
IntFOLD5 COMA multi3 TS1 4y9jA |
CERT: 1.485E-9 |
0.5976 |
 |
|
|
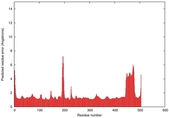
IntFOLD5 COMA multi2 TS1 4y9jA |
CERT: 1.485E-9 |
0.5976 |
 |
|
|
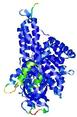
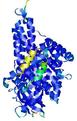
IntFOLD5 MUSTER multi7 TS1 4y9jA 4y9jA1 |
CERT: 1.52E-9 |
0.5971 |
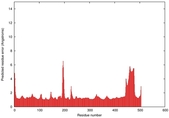 |
|
|
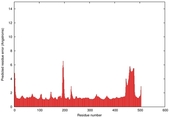
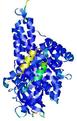
IntFOLD5 MUSTER multi3 TS1 4y9jA 4y9jA1 |
CERT: 1.52E-9 |
0.5971 |
 |
|
|
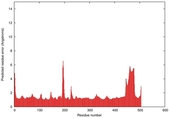
IntFOLD5 MUSTER multi1 TS1 4y9jA 4y9jA1 |
CERT: 1.52E-9 |
0.5971 |
 |
|
|
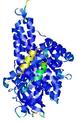
IntFOLD5 COMA multi5 TS1 4y9jA 3u33A |
CERT: 1.528E-9 |
0.5970 |
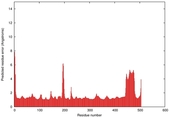 |
|
|
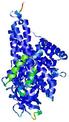
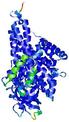
IntFOLD5 COMA multi1 TS1 4y9jA 3u33A |
CERT: 1.528E-9 |
0.5970 |
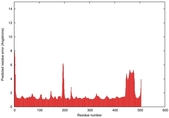 |
|
|
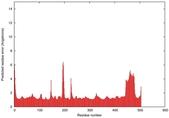
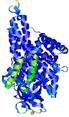
IntFOLD5 wPPAS multi5 TS1 4y9jA 5ez3A |
CERT: 1.529E-9 |
0.5970 |
 |
|
|
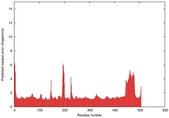
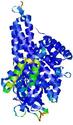
IntFOLD5 wdPPAS multi5 TS1 4y9jA 5ez3A |
CERT: 1.581E-9 |
0.5964 |
 |
|
|
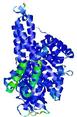
IntFOLD5 PPAS multi5 TS1 4y9jA 5ez3A |
CERT: 1.605E-9 |
0.5961 |
 |
|
|
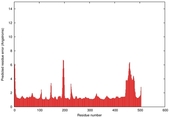
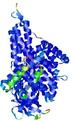
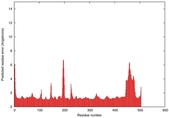
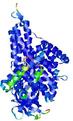
IntFOLD5 SPARKSX multi5 TS1 4y9jA 3djlA |
CERT: 1.639E-9 |
0.5957 |
 |
|
|
IntFOLD5 SPARKSX multi1 TS1 4y9jA 3djlA |
CERT: 1.639E-9 |
0.5957 |
 |
|
|
IntFOLD5 sp3 multi8 TS1 4y9jA |
CERT: 1.662E-9 |
0.5954 |
 |

|
|
IntFOLD5 sp3 multi7 TS1 4y9jA |
CERT: 1.662E-9 |
0.5954 |
 |

|
|
IntFOLD5 sp3 multi6 TS1 4y9jA |
CERT: 1.662E-9 |
0.5954 |
 |

|
|
IntFOLD5 sp3 multi4 TS1 4y9jA |
CERT: 1.662E-9 |
0.5954 |
 |

|
|
IntFOLD5 sp3 multi3 TS1 4y9jA |
CERT: 1.662E-9 |
0.5954 |
 |

|
|
IntFOLD5 sp3 multi2 TS1 4y9jA |
CERT: 1.662E-9 |
0.5954 |
 |

|
|
IntFOLD5 HHpred TS1 3djlA 4y9jA 5ez3A |
CERT: 1.691E-9 |
0.5951 |
 |

|
|
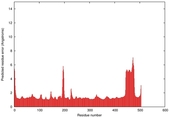
IntFOLD5 dPPAS2 multi8 TS1 4y9jA |
CERT: 1.755E-9 |
0.5943 |
 |

|
|
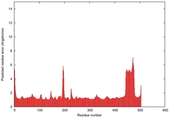
IntFOLD5 dPPAS2 multi6 TS1 4y9jA |
CERT: 1.755E-9 |
0.5943 |
 |

|
|
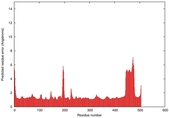
IntFOLD5 dPPAS2 multi4 TS1 4y9jA |
CERT: 1.755E-9 |
0.5943 |
 |

|
|
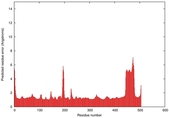
IntFOLD5 dPPAS2 multi2 TS1 4y9jA |
CERT: 1.755E-9 |
0.5943 |
 |

|
|
IntFOLD5 dPPAS multi4 TS1 4y9jA |
CERT: 1.81E-9 |
0.5938 |
 |

|
|
IntFOLD5 dPPAS multi2 TS1 4y9jA |
CERT: 1.81E-9 |
0.5938 |
 |

|
|
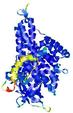
IntFOLD5 wdPPAS multi1 TS1 4y9jA 4y9jA1 |
CERT: 1.841E-9 |
0.5934 |
 |
|
|
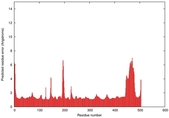
IntFOLD5 HHsearch multi5 TS1 4y9jA 5ez3A |
CERT: 1.843E-9 |
0.5934 |
 |
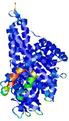
|
|
IntFOLD5 wdPPAS multi8 TS1 4y9jA |
CERT: 1.844E-9 |
0.5934 |
 |
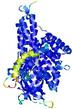
|
|
IntFOLD5 wdPPAS multi6 TS1 4y9jA |
CERT: 1.844E-9 |
0.5934 |
 |
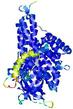
|
|
IntFOLD5 wdPPAS multi4 TS1 4y9jA |
CERT: 1.844E-9 |
0.5934 |
 |
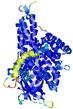
|
|
IntFOLD5 wdPPAS multi2 TS1 4y9jA |
CERT: 1.844E-9 |
0.5934 |
 |
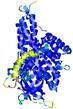
|
|
IntFOLD5 Env-PPAS multi7 TS1 4y9jA 4y9jA1 |
CERT: 1.866E-9 |
0.5932 |
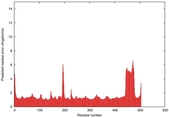 |
|
|
IntFOLD5 SPARKSX multi8 TS1 4y9jA |
CERT: 1.87E-9 |
0.5931 |
 |

|
|
IntFOLD5 SPARKSX multi7 TS1 4y9jA |
CERT: 1.87E-9 |
0.5931 |
 |

|
|
IntFOLD5 SPARKSX multi6 TS1 4y9jA |
CERT: 1.87E-9 |
0.5931 |
 |

|
|
IntFOLD5 SPARKSX multi4 TS1 4y9jA |
CERT: 1.87E-9 |
0.5931 |
 |

|
|
IntFOLD5 SPARKSX multi3 TS1 4y9jA |
CERT: 1.87E-9 |
0.5931 |
 |

|
|
IntFOLD5 SPARKSX multi2 TS1 4y9jA |
CERT: 1.87E-9 |
0.5931 |
 |

|
|
IntFOLD5 HHpred TS2 3djlA 4y9jA 5ez3A |
CERT: 1.881E-9 |
0.5930 |
 |

|
|
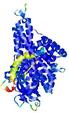
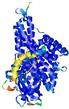
IntFOLD5 Env-PPAS multi8 TS1 4y9jA |
CERT: 1.902E-9 |
0.5928 |
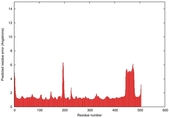 |
|
|
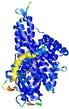
IntFOLD5 Env-PPAS multi6 TS1 4y9jA |
CERT: 1.902E-9 |
0.5928 |
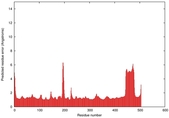 |
|
|
IntFOLD5 LOMETS multi3 TS1 4y9jA 4y9jA1 |
CERT: 1.908E-9 |
0.5927 |
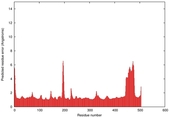 |

|
|
IntFOLD5 wPPAS multi8 TS1 4y9jA |
CERT: 1.937E-9 |
0.5924 |
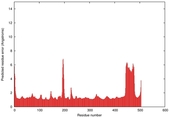 |
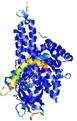
|
|
IntFOLD5 wPPAS multi6 TS1 4y9jA |
CERT: 1.937E-9 |
0.5924 |
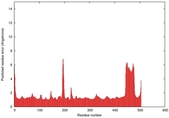 |
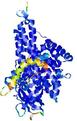
|
|
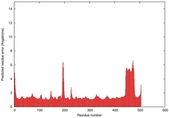
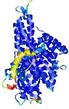
IntFOLD5 Env-PPAS multi4 TS1 4y9jA1 4y9jA |
CERT: 1.945E-9 |
0.5924 |
 |
|
|
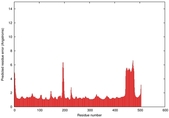
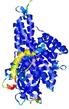
IntFOLD5 Env-PPAS multi1 TS1 4y9jA1 4y9jA |
CERT: 1.945E-9 |
0.5924 |
 |
|
|
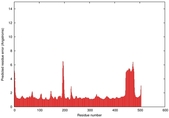
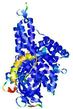
IntFOLD5 PPAS multi4 TS1 4y9jA1 4y9jA |
CERT: 1.951E-9 |
0.5923 |
 |
|
|
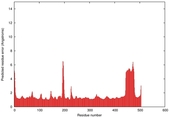
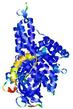
IntFOLD5 PPAS multi1 TS1 4y9jA1 4y9jA |
CERT: 1.951E-9 |
0.5923 |
 |
|
|
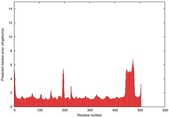
IntFOLD5 LOMETS multi1 TS1 4y9jA |
CERT: 1.958E-9 |
0.5922 |
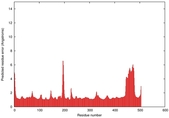 |
|
|
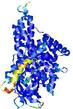
IntFOLD5 dPPAS multi1 TS1 4y9jA 4y9jA1 |
CERT: 1.96E-9 |
0.5922 |
 |
|
|
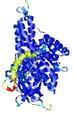
IntFOLD5 dPPAS2 multi1 TS1 4y9jA 4y9jA1 |
CERT: 1.993E-9 |
0.5919 |
 |

|
|
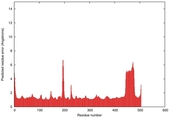
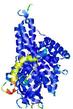
IntFOLD5 PPAS multi8 TS1 4y9jA |
CERT: 2.04E-9 |
0.5915 |
 |
|
|
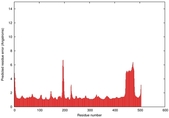
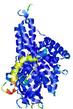
IntFOLD5 PPAS multi6 TS1 4y9jA |
CERT: 2.04E-9 |
0.5915 |
 |
|
|
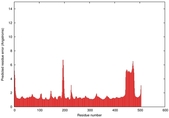
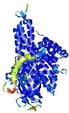
IntFOLD5 PPAS multi7 TS1 4y9jA 4y9jA1 |
CERT: 2.126E-9 |
0.5907 |
 |
|
|
IntFOLD5 dPPAS multi5 TS1 5ez3A 4y9jA |
CERT: 2.133E-9 |
0.5906 |
 |

|
|
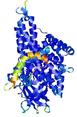
IntFOLD5 wPPAS multi7 TS1 4y9jA 4y9jA1 |
CERT: 2.163E-9 |
0.5903 |
 |
|
|
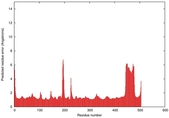
IntFOLD5 wPPAS multi4 TS1 4y9jA1 4y9jA |
CERT: 2.169E-9 |
0.5903 |
 |
|
|
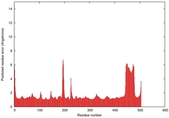
IntFOLD5 wPPAS multi1 TS1 4y9jA1 4y9jA |
CERT: 2.169E-9 |
0.5903 |
 |
|
|
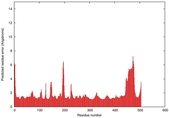
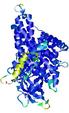
IntFOLD5 IT5 TS1 |
CERT: 4.215E-9 |
0.5777 |
 |
|
|
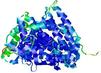
IntFOLD5 IntFOLDTS140 multi2 TS1 5ez3A |
CERT: 1.859E-8 |
0.5504 |
 |
|
|
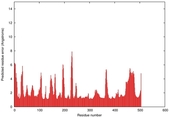
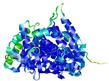
IntFOLD5 dPPAS multi8 TS1 5ez3A |
CERT: 2.665E-8 |
0.5440 |
 |
|
|
IntFOLD5 dPPAS multi6 TS1 5ez3A |
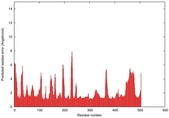
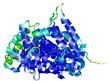
CERT: 2.665E-8 |
0.5440 |
 |
|
|
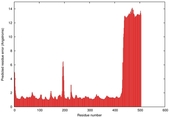
IntFOLD5 wPPAS multi3 TS1 4y9jA1 |
CERT: 4.19E-8 |
0.5361 |
 |

|
|
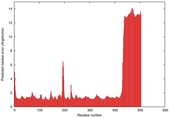
IntFOLD5 wPPAS multi2 TS1 4y9jA1 |
CERT: 4.19E-8 |
0.5361 |
 |

|
|
IntFOLD5 PPAS multi3 TS1 4y9jA1 |
CERT: 4.431E-8 |
0.5351 |
 |

|
|
IntFOLD5 PPAS multi2 TS1 4y9jA1 |
CERT: 4.431E-8 |
0.5351 |
 |

|
|
IntFOLD5 Env-PPAS multi3 TS1 4y9jA1 |
CERT: 4.431E-8 |
0.5351 |
 |

|
|
IntFOLD5 Env-PPAS multi2 TS1 4y9jA1 |
CERT: 4.431E-8 |
0.5351 |
 |

|