The IntFOLD Server Results Page (Version 5.0)
Please cite the relevant papers:McGuffin, L.J., Adiyaman, R., Maghrabi, A.H.A., Shuid, A.N., Brackenridge, D.A., Nealon, J.O. & Philomina, L.S. (2019) IntFOLD: an integrated web resource for high performance protein structure and function prediction. Nucleic Acids Research, 47, W408-W413. DOI PubMed
McGuffin, L.J., Shuid, A.M., Kempster, R., Maghrabi, A.H.A., Nealon J.O., Salehe, B.R., Atkins, J.D. & Roche, D.B. (2017) Accurate Template Based Modelling in CASP12 using the IntFOLD4-TS, ModFOLD6 and ReFOLD methods. Proteins: Structure, Function, and Bioinformatics, 86 Suppl 1, 335-344, doi: 10.1002/prot.25360. PubMed
Links to graphical output:
- Top 5 3D models
- Disorder prediction
- Domain boundary prediction
- Binding site prediction
- Full model quality assessment results
Download machine readable results in CASP format:
- TS (Tertiary Structure Prediction)
- DR (Disorder Prediction)
- DP (Domain Prediction)
- FN (Binding Site Prediction)
- QA (Model Quality Prediction)
- Make a compressed file containing all generated 3D models (models are in PDB format with predicted per-residue errors in the B-factor column)
Results will be available for 21 days after job completion (subject to server capacity)
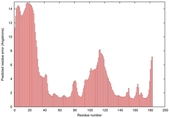
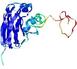
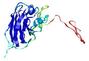
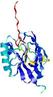
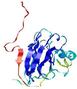
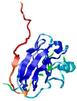
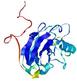
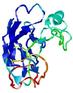
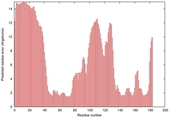
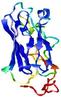
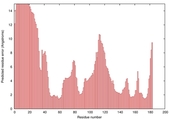
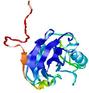
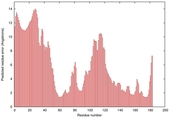
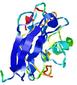
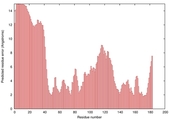
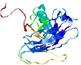
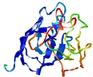
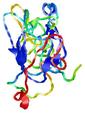
| Tertiary Structure Predictions: Top 5 multi-template 3D models for T0949 | Help | ||||
| Model ID and PDBsum links for all templates used | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | 3D map of model quality |
|
IntFOLD5 IT5 TS1 |
CERT: 2.047E-5 |
0.4383 |
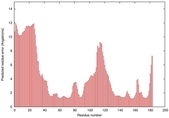 |
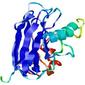
|
|
IntFOLD5 MUSTER multi7 TS1 1ov8A 3ay2A |
CERT: 6.171E-5 |
0.4229 |
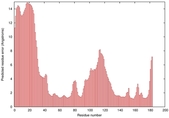 |
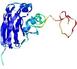
|
|
IntFOLD5 MUSTER multi5 TS1 1ov8A 3ay2A |
CERT: 6.171E-5 |
0.4229 |
 |
|
|
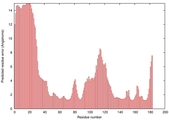
IntFOLD5 IntFOLDTS140 multi1 TS1 1qhqA 1cuoA |
CERT: 9.296E-5 |
0.4173 |
 |

|
|
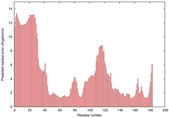
IntFOLD5 IT5 TS3 |
CERT: 1.021E-4 |
0.4160 |
 |
|
McGuffin, L. J. (2008) Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics, 24, 1789-1804. PubMed
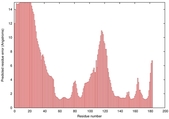
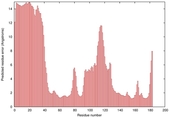
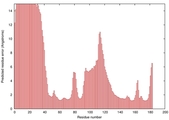
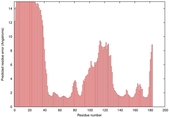
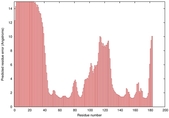
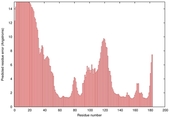
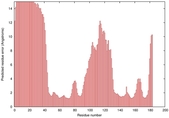
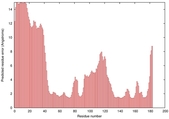
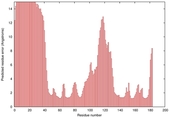
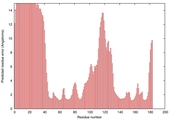
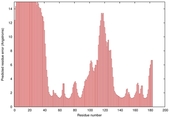
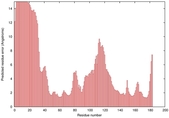
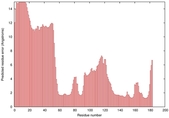
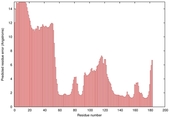
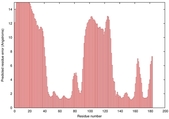
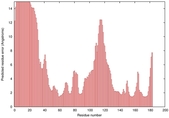
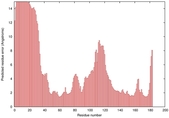
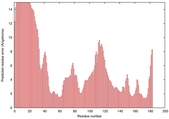
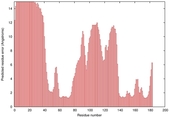
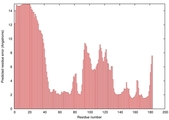
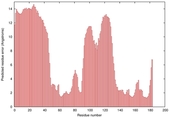
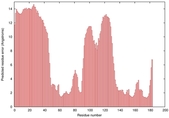
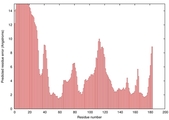
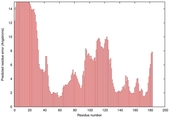
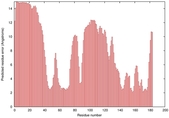
| Disorder prediction for T0949 | Help | |||
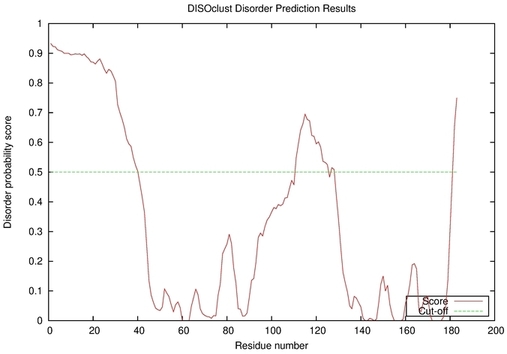
| |||
| Click image to download plot in PostScript format. |
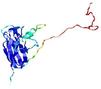
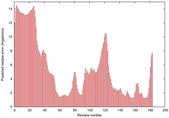
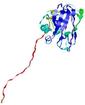
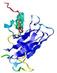
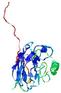
| Domain boundary prediction for T0949 | Help | |||

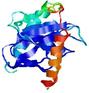
| |||
| Click image to download model or view prediction using Jmol. The model above is coloured according to the predicted domains. |
Roche, D. B., Tetchner, S. J. & McGuffin, L. J. (2011) FunFOLD: an improved automated method for the prediction of ligand binding residues using 3D models of proteins. BMC Bioinformatics, 12, 160. PubMed
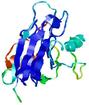
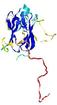
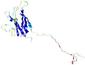
| Ligand binding residues prediction for T0949 | Help | |||
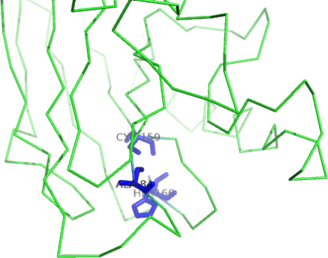
| |||
| Click image to download model or view prediction using Jmol. Predicted ligand binding residues are shown as blue sticks in the image above. |
Maghrabi, A.H.A. & McGuffin L.J. (2017) ModFOLD6: an accurate web server for the global and local quality estimation of 3D models of proteins. Nucleic Acids Res., 45, W416-W421, doi: 10.1093/nar/gkx332. PubMed
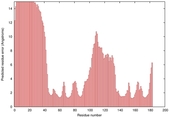
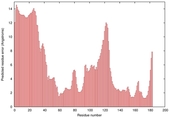
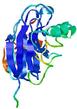
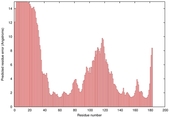
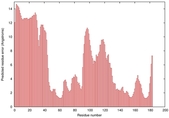
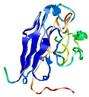
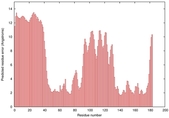
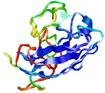
| Full model quality assessment results for T0949 | Help | ||||
| Model ID and PDBsum links for all templates used | Confidence and P-value | Global model quality score | Local model quality plot (click images to download plots) | 3D map of model quality |
|
IntFOLD5 IT5 TS1 |
CERT: 2.047E-5 |
0.4383 |
 |

|
|
IntFOLD5 MUSTER multi7 TS1 1ov8A 3ay2A |
CERT: 6.171E-5 |
0.4229 |
 |

|
|
IntFOLD5 MUSTER multi5 TS1 1ov8A 3ay2A |
CERT: 6.171E-5 |
0.4229 |
 |

|
|
IntFOLD5 IntFOLDTS140 multi1 TS1 1qhqA 1cuoA |
CERT: 9.296E-5 |
0.4173 |
 |

|
|
IntFOLD5 IT5 TS3 |
CERT: 1.021E-4 |
0.4160 |
 |

|
|
IntFOLD5 PPAS multi1 TS1 1ov8A 2aanA |
CERT: 1.093E-4 |
0.4151 |
 |

|
|
IntFOLD5 wPPAS multi1 TS1 1ov8A 2aanA |
CERT: 1.245E-4 |
0.4133 |
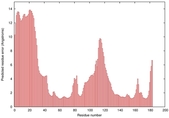 |

|
|
IntFOLD5 wMUSTER multi5 TS1 1ov8A 2aanA |
CERT: 1.269E-4 |
0.4131 |
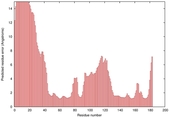 |
|
|
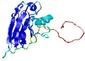
IntFOLD5 wMUSTER multi1 TS1 1ov8A 2aanA |
CERT: 1.269E-4 |
0.4131 |
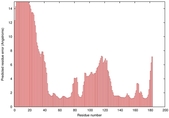 |
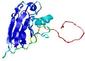
|
|
IntFOLD5 IntFOLDTS140 multi4 TS1 1qhqA 1cuoA 3tasA 1fwxA2 |
CERT: 1.281E-4 |
0.4129 |
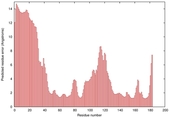 |
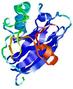
|
|
IntFOLD5 MUSTER multi6 TS1 1ov8A |
CERT: 1.291E-4 |
0.4128 |
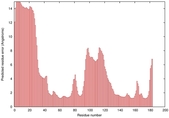 |

|
|
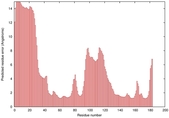
IntFOLD5 MUSTER multi2 TS1 1ov8A |
CERT: 1.291E-4 |
0.4128 |
 |

|
|
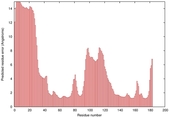
IntFOLD5 LOMETS multi2 TS1 1ov8A |
CERT: 1.291E-4 |
0.4128 |
 |

|
|
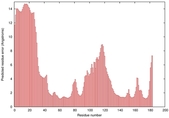
IntFOLD5 MUSTER multi1 TS1 1ov8A 2aanA |
CERT: 1.332E-4 |
0.4124 |
 |
|
|
IntFOLD5 wPPAS multi7 TS1 1ov8A |
CERT: 1.394E-4 |
0.4118 |
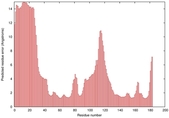 |
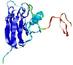
|
|
IntFOLD5 wPPAS multi6 TS1 1ov8A |
CERT: 1.394E-4 |
0.4118 |
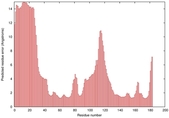 |
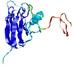
|
|
IntFOLD5 wPPAS multi3 TS1 1ov8A |
CERT: 1.394E-4 |
0.4118 |
 |
|
|
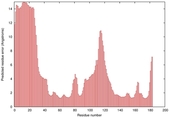
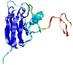
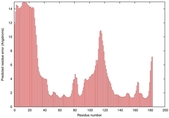
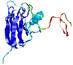
IntFOLD5 wPPAS multi2 TS1 1ov8A |
CERT: 1.394E-4 |
0.4118 |
 |
|
|
IntFOLD5 wdPPAS multi2 TS1 1ov8A |
CERT: 1.394E-4 |
0.4118 |
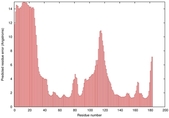 |
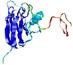
|
|
IntFOLD5 wMUSTER multi7 TS1 1ov8A 3ay2A 4hz1A |
CERT: 1.405E-4 |
0.4117 |
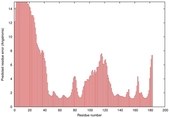 |
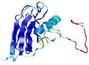
|
|
IntFOLD5 wMUSTER multi3 TS1 1ov8A 3ay2A 4hz1A |
CERT: 1.405E-4 |
0.4117 |
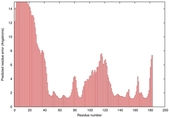 |
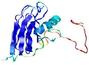
|
|
IntFOLD5 IT5 TS2 |
CERT: 1.458E-4 |
0.4112 |
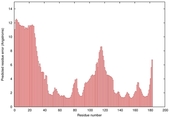 |
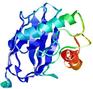
|
|
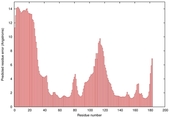
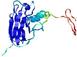
IntFOLD5 LOMETS multi3 TS1 1ov8A |
CERT: 1.477E-4 |
0.4110 |
 |
|
|
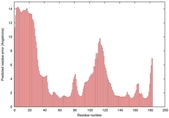
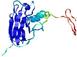
IntFOLD5 LOMETS multi1 TS1 1ov8A |
CERT: 1.477E-4 |
0.4110 |
 |
|
|
IntFOLD5 wdPPAS multi5 TS1 1cuoA 1ov8A |
CERT: 1.544E-4 |
0.4104 |
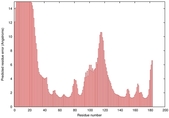 |
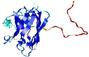
|
|
IntFOLD5 wPPAS multi5 TS1 1ov8A 1cuoA |
CERT: 1.561E-4 |
0.4103 |
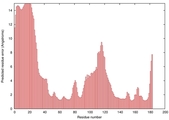 |
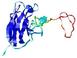
|
|
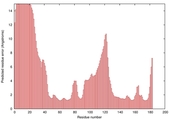
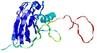
IntFOLD5 Env-PPAS multi1 TS1 1ov8A 2aanA |
CERT: 1.567E-4 |
0.4102 |
 |
|
|
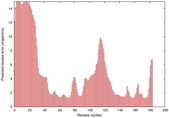
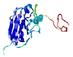
IntFOLD5 PPAS multi5 TS1 1ov8A 3ay2A |
CERT: 1.575E-4 |
0.4102 |
 |
|
|
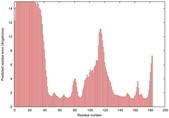
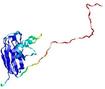
IntFOLD5 CNFsearch multi7 TS1 1ov8A 4bwwA |
CERT: 1.596E-4 |
0.4100 |
 |
|
|
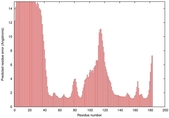
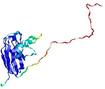
IntFOLD5 CNFsearch multi5 TS1 1ov8A 4bwwA |
CERT: 1.596E-4 |
0.4100 |
 |
|
|
IntFOLD5 CNFsearch multi3 TS1 1ov8A 4bwwA |
CERT: 1.596E-4 |
0.4100 |
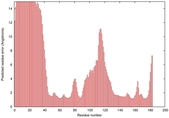 |
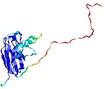
|
|
IntFOLD5 dPPAS2 multi5 TS1 1ov8A 3ay2A |
CERT: 1.664E-4 |
0.4094 |
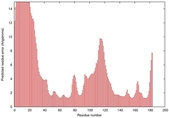 |

|
|
IntFOLD5 LOMETS multi4 TS1 1ov8A |
CERT: 1.68E-4 |
0.4093 |
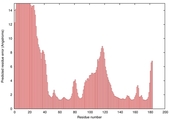 |
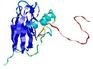
|
|
IntFOLD5 COMA multi5 TS1 1qhqA 2ccwA |
CERT: 1.683E-4 |
0.4093 |
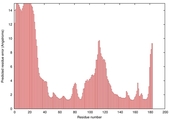 |

|
|
IntFOLD5 HHsearch multi4 TS1 5tk2A 1nwpA 3ay2A 2aanA 1qhqA |
CERT: 1.855E-4 |
0.4080 |
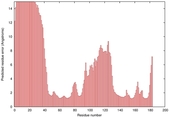 |

|
|
IntFOLD5 SPARKSX multi4 TS1 1qhqA |
CERT: 1.879E-4 |
0.4078 |
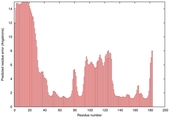 |
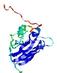
|
|
IntFOLD5 SPARKSX multi2 TS1 1qhqA |
CERT: 1.879E-4 |
0.4078 |
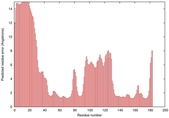 |
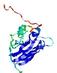
|
|
IntFOLD5 SPARKSX multi8 TS1 1cuoA 1qhqA |
CERT: 1.911E-4 |
0.4076 |
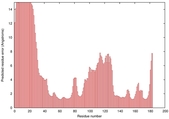 |
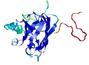
|
|
IntFOLD5 dPPAS2 multi1 TS1 1ov8A 2aanA |
CERT: 1.961E-4 |
0.4073 |
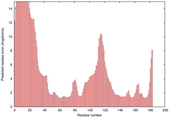 |

|
|
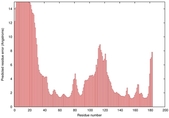
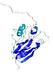
IntFOLD5 IntFOLDTS140 multi3 TS1 1qhqA |
CERT: 2.084E-4 |
0.4064 |
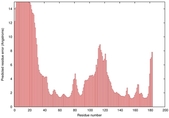 |
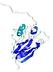
|
|
IntFOLD5 IntFOLDTS140 multi2 TS1 1qhqA |
CERT: 2.084E-4 |
0.4064 |
 |
|
|
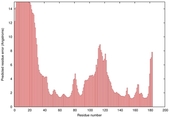
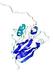
IntFOLD5 COMA multi8 TS1 1qhqA |
CERT: 2.084E-4 |
0.4064 |
 |
|
|
IntFOLD5 COMA multi7 TS1 1qhqA |
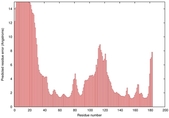
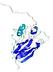
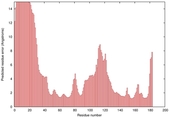
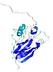
CERT: 2.084E-4 |
0.4064 |
 |
|
|
IntFOLD5 COMA multi6 TS1 1qhqA |
CERT: 2.084E-4 |
0.4064 |
 |
|
|
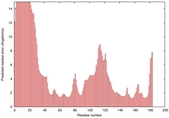
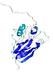
IntFOLD5 COMA multi4 TS1 1qhqA |
CERT: 2.084E-4 |
0.4064 |
 |
|
|
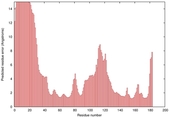
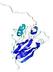
IntFOLD5 COMA multi3 TS1 1qhqA |
CERT: 2.084E-4 |
0.4064 |
 |
|
|
IntFOLD5 COMA multi2 TS1 1qhqA |
CERT: 2.084E-4 |
0.4064 |
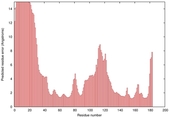 |
|
|
IntFOLD5 wdPPAS multi3 TS1 1ov8A 1cuoA |
CERT: 2.091E-4 |
0.4064 |
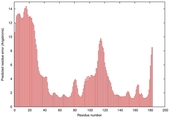 |
|
|
IntFOLD5 PPAS multi7 TS1 1ov8A |
CERT: 2.1E-4 |
0.4063 |
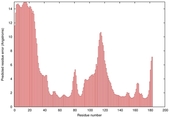 |
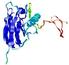
|
|
IntFOLD5 PPAS multi6 TS1 1ov8A |
CERT: 2.1E-4 |
0.4063 |
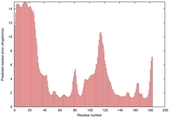 |
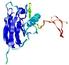
|
|
IntFOLD5 PPAS multi3 TS1 1ov8A |
CERT: 2.1E-4 |
0.4063 |
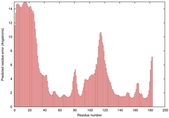 |
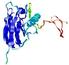
|
|
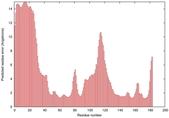
IntFOLD5 PPAS multi2 TS1 1ov8A |
CERT: 2.1E-4 |
0.4063 |
 |
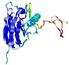
|
|
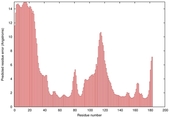
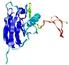
IntFOLD5 dPPAS multi7 TS1 1ov8A |
CERT: 2.1E-4 |
0.4063 |
 |
|
|
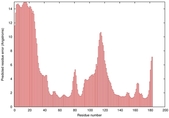
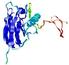
IntFOLD5 dPPAS multi6 TS1 1ov8A |
CERT: 2.1E-4 |
0.4063 |
 |
|
|
IntFOLD5 dPPAS2 multi7 TS1 1ov8A |
CERT: 2.1E-4 |
0.4063 |
 |
|
|
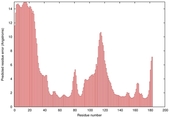
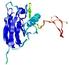
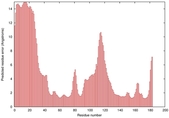
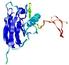
IntFOLD5 dPPAS2 multi6 TS1 1ov8A |
CERT: 2.1E-4 |
0.4063 |
 |
|
|
IntFOLD5 dPPAS2 multi3 TS1 1ov8A |
CERT: 2.1E-4 |
0.4063 |
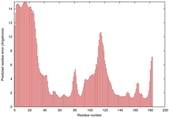 |
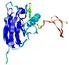
|
|
IntFOLD5 dPPAS2 multi2 TS1 1ov8A |
CERT: 2.1E-4 |
0.4063 |
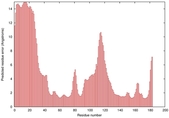 |
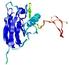
|
|
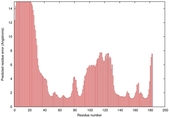
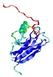
IntFOLD5 SPARKSX multi3 TS1 1qhqA 2aanA 1cuoA |
CERT: 2.141E-4 |
0.4061 |
 |
|
|
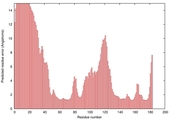
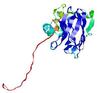
IntFOLD5 HHpred TS1 1cuoA 2wtpA 2aanA 2dv6A 5tk2A |
CERT: 2.162E-4 |
0.4060 |
 |
|
|
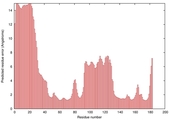
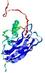
IntFOLD5 SPARKSX multi1 TS1 1qhqA 2aanA |
CERT: 2.185E-4 |
0.4058 |
 |
|
|
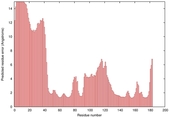
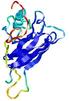
IntFOLD5 spk2 multi7 TS1 1qhqA |
CERT: 2.355E-4 |
0.4048 |
 |
|
|
IntFOLD5 spk2 multi6 TS1 1qhqA |
CERT: 2.355E-4 |
0.4048 |
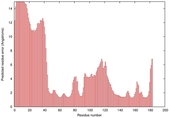 |
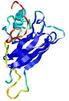
|
|
IntFOLD5 Env-PPAS multi6 TS1 1ov8A |
CERT: 2.417E-4 |
0.4045 |
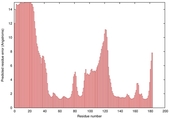 |
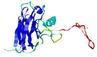
|
|
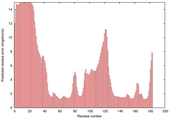
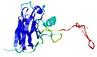
IntFOLD5 Env-PPAS multi2 TS1 1ov8A |
CERT: 2.417E-4 |
0.4045 |
 |
|
|
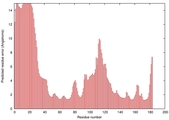
IntFOLD5 COMA multi1 TS1 1qhqA 2ft6A |
CERT: 2.513E-4 |
0.4040 |
 |

|
|
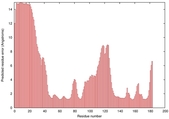
IntFOLD5 Env-PPAS multi7 TS1 1ov8A 3ay2A |
CERT: 2.576E-4 |
0.4037 |
 |

|
|
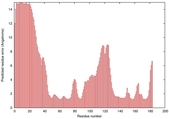
IntFOLD5 Env-PPAS multi5 TS1 1ov8A 3ay2A |
CERT: 2.576E-4 |
0.4037 |
 |

|
|
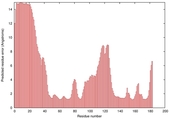
IntFOLD5 Env-PPAS multi3 TS1 1ov8A 3ay2A |
CERT: 2.576E-4 |
0.4037 |
 |

|
|
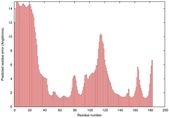
IntFOLD5 dPPAS multi5 TS1 1ov8A 3ay2A |
CERT: 2.594E-4 |
0.4036 |
 |
|
|
IntFOLD5 dPPAS multi1 TS1 1ov8A 2aanA |
CERT: 2.777E-4 |
0.4027 |
 |

|
|
IntFOLD5 CNFsearch multi8 TS1 1ov8A 3tasA |
CERT: 2.862E-4 |
0.4023 |
 |

|
|
IntFOLD5 CNFsearch multi6 TS1 1ov8A |
CERT: 2.914E-4 |
0.4020 |
 |
|
|
IntFOLD5 CNFsearch multi2 TS1 1ov8A |
CERT: 2.914E-4 |
0.4020 |
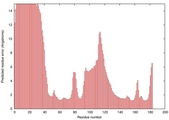 |
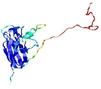
|
|
IntFOLD5 IntFOLDTS60 multi3 TS1 2ccwA 1qhqA |
CERT: 3.004E-4 |
0.4016 |
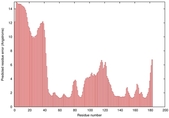 |
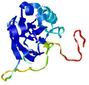
|
|
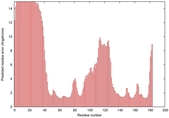
IntFOLD5 HHsearch multi8 TS1 1cuoA 2aanA 1qhqA |
CERT: 3.066E-4 |
0.4014 |
 |

|
|
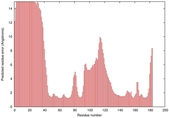
IntFOLD5 CNFsearch multi1 TS1 1ov8A 4hcfA |
CERT: 3.264E-4 |
0.4006 |
 |

|
|
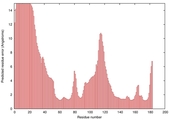
IntFOLD5 dPPAS multi2 TS1 1ov8A |
CERT: 3.904E-4 |
0.3982 |
 |
|
|
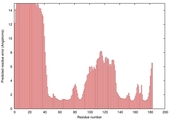
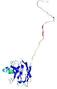
IntFOLD5 SPARKSX multi5 TS1 1cuoA 2aanA |
CERT: 3.914E-4 |
0.3982 |
 |
|
|
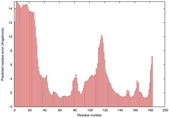
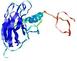
IntFOLD5 wdPPAS multi1 TS1 1ov8A 2aanA |
CERT: 3.928E-4 |
0.3982 |
 |
|
|
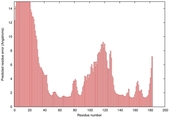
IntFOLD5 CNFsearch multi4 TS1 1ov8A 1ibyA 4f2fA 3tasA |
CERT: 4.301E-4 |
0.3970 |
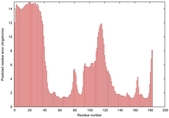 |
|
|
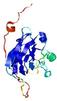
IntFOLD5 MUSTER multi3 TS1 1ov8A 2aanA 3ay2A |
CERT: 4.578E-4 |
0.3962 |
 |

|
|
IntFOLD5 dPPAS multi3 TS1 1ov8A 3ay2A 1cuoA |
CERT: 5.051E-4 |
0.3949 |
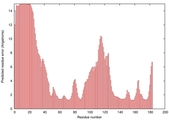 |
|
|
IntFOLD5 HHsearch multi3 TS1 5tk2A 1nwpA 2iaaC 2ccwA 2aanA |
CERT: 5.337E-4 |
0.3942 |
 |

|
|
IntFOLD5 Env-PPAS multi8 TS1 1ov8A 1fwxA2 |
CERT: 5.861E-4 |
0.3930 |
 |
|
|
IntFOLD5 wMUSTER multi6 TS1 1ov8A |
CERT: 6.684E-4 |
0.3913 |
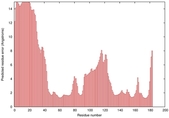 |
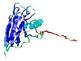
|
|
IntFOLD5 wMUSTER multi2 TS1 1ov8A |
CERT: 6.684E-4 |
0.3913 |
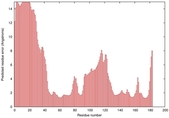 |
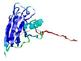
|
|
IntFOLD5 HHsearch multi7 TS1 1cuoA 2ccwA 2aanA |
CERT: 6.885E-4 |
0.3910 |
 |

|
|
IntFOLD5 HHpred TS3 1cuoA 2wtpA 2aanA 2dv6A 5tk2A |
CERT: 7.309E-4 |
0.3902 |
 |
|
|
IntFOLD5 HHsearch multi1 TS1 5tk2A 1nwpA |
CERT: 7.742E-4 |
0.3895 |
 |

|
|
IntFOLD5 spk2 multi5 TS1 1qhqA 4f2eA |
CERT: 7.772E-4 |
0.3894 |
 |
|
|
IntFOLD5 HHsearch multi5 TS1 1cuoA 2ccwA |
CERT: 9.08E-4 |
0.3875 |
 |

|
|
IntFOLD5 IntFOLDTS60 multi1 TS1 2ccwA 1cuoA |
CERT: 9.499E-4 |
0.3869 |
 |

|
|
IntFOLD5 spk2 multi3 TS1 4f2eA 1qhqA |
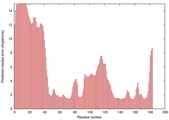
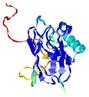
CERT: 9.743E-4 |
0.3866 |
 |
|
|
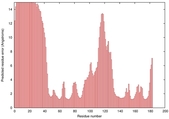
IntFOLD5 HHsearch multi6 TS1 1cuoA |
HIGH: 1.136E-3 |
0.3846 |
 |

|
|
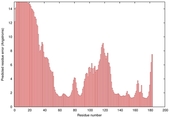
IntFOLD5 HHpred TS2 1cuoA 2wtpA 2aanA 2dv6A 5tk2A |
HIGH: 1.197E-3 |
0.3840 |
 |
|
|
IntFOLD5 IntFOLDTS60 multi2 TS1 2ccwA |
HIGH: 1.273E-3 |
0.3832 |
 |

|
|
IntFOLD5 wPPAS multi4 TS1 1ov8A 6hbeA2 2w88A 2dv6A1 |
HIGH: 1.529E-3 |
0.3809 |
 |
|
|
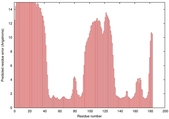
IntFOLD5 HHsearch multi2 TS1 5tk2A |
HIGH: 2.155E-3 |
0.3767 |
 |

|
|
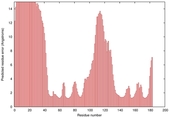
IntFOLD5 wdPPAS multi7 TS1 1cuoA |
HIGH: 2.23E-3 |
0.3763 |
 |

|
|
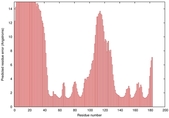
IntFOLD5 wdPPAS multi6 TS1 1cuoA |
HIGH: 2.23E-3 |
0.3763 |
 |

|
|
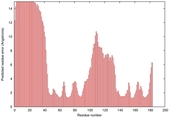
IntFOLD5 SPARKSX multi7 TS1 1cuoA |
HIGH: 2.455E-3 |
0.3751 |
 |

|
|
IntFOLD5 SPARKSX multi6 TS1 1cuoA |
HIGH: 2.455E-3 |
0.3751 |
 |

|
|
IntFOLD5 Env-PPAS multi4 TS1 1ov8A 6hbeA2 1qniA2 |
HIGH: 2.466E-3 |
0.3751 |
 |
|
|
IntFOLD5 wdPPAS multi8 TS1 1cuoA 1ov8A 2h3xC 2dv6A1 6hbeA2 |
HIGH: 3.468E-3 |
0.3709 |
 |
|
|
IntFOLD5 sp3 multi7 TS1 1qhqA |
HIGH: 4.051E-3 |
0.3691 |
 |
|
|
IntFOLD5 sp3 multi6 TS1 1qhqA |
HIGH: 4.051E-3 |
0.3691 |
 |
|
|
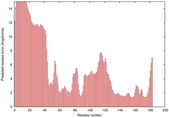
IntFOLD5 sp3 multi3 TS1 1aozA1 1qhqA |
HIGH: 5.13E-3 |
0.3662 |
 |
|
|
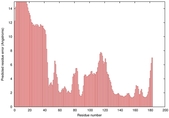
IntFOLD5 sp3 multi1 TS1 1aozA1 1qhqA |
HIGH: 5.13E-3 |
0.3662 |
 |
|
|
IntFOLD5 spk2 multi2 TS1 4f2eA |
HIGH: 5.163E-3 |
0.3662 |
 |
|
|
IntFOLD5 wdPPAS multi4 TS1 1ov8A 6hbeA2 2w88A 2dv6A1 |
HIGH: 5.687E-3 |
0.3650 |
 |
|
|
IntFOLD5 wPPAS multi8 TS1 1ov8A 2dv6A1 6hbeA2 |
HIGH: 6.958E-3 |
0.3626 |
 |
|
|
IntFOLD5 PPAS multi4 TS1 1ov8A 6hbeA2 2w88A |
MEDIUM: 1.109E-2 |
0.3572 |
 |
|
|
IntFOLD5 sp3 multi2 TS1 1aozA1 |
MEDIUM: 1.544E-2 |
0.3533 |
 |
|
|
IntFOLD5 IntFOLDTS60 multi4 TS1 2ccwA 1cuoA 2aanA 1qhqA 3tasA 1gw0A1 1aozA1 1j9qA1 |
MEDIUM: 2.034E-2 |
0.3502 |
 |
|
|
IntFOLD5 spk2 multi1 TS1 4f2eA 1gw0A1 |
MEDIUM: 2.08E-2 |
0.3499 |
 |
|
|
IntFOLD5 dPPAS2 multi4 TS1 1ov8A 6hbeA2 2w88A |
MEDIUM: 2.649E-2 |
0.3469 |
 |
|
|
IntFOLD5 dPPAS multi8 TS1 1ov8A 3gyrL2 |
MEDIUM: 2.791E-2 |
0.3462 |
 |
|
|
IntFOLD5 sp3 multi5 TS1 1qhqA 1kbvA1 |
MEDIUM: 4.896E-2 |
0.3369 |
 |
|
|
IntFOLD5 wMUSTER multi8 TS1 1ov8A 3gyrL2 |
LOW: 5.358E-2 |
0.3350 |
 |
|
|
IntFOLD5 wMUSTER multi4 TS1 1ov8A 3gyrL2 |
LOW: 5.358E-2 |
0.3350 |
 |
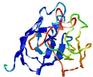
|
|
IntFOLD5 dPPAS2 multi8 TS1 1ov8A 2w88A 3c75A 6hbeA2 |
LOW: 5.641E-2 |
0.3339 |
 |

|
|
IntFOLD5 PPAS multi8 TS1 1ov8A 2gimA 3c75A 6hbeA2 |
LOW: 6.051E-2 |
0.3323 |
 |
|
|
IntFOLD5 MUSTER multi8 TS1 1ov8A 1cuoA 3c75A 2w88A 3gyrL2 |
LOW: 6.975E-2 |
0.3288 |
 |
|
|
IntFOLD5 spk2 multi8 TS1 1qhqA 4f2eA 1ibyA 1gw0A1 1aozA1 1j9qA1 |
LOW: 7.81E-2 |
0.3257 |
 |
|
|
IntFOLD5 sp3 multi4 TS1 1aozA1 1qhqA 1gw0A1 1j9qA1 1gskA |
UNCERT: 1.033E-1 |
0.3171 |
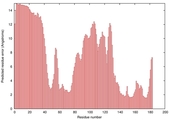 |
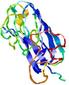
|
|
IntFOLD5 sp3 multi8 TS1 1qhqA 1kbvA1 1gw0A1 1gskA |
UNCERT: 1.187E-1 |
0.3126 |
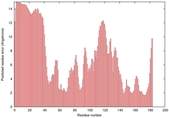 |
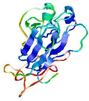
|
|
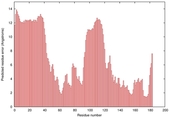
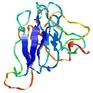
IntFOLD5 dPPAS multi4 TS1 1ov8A 6hbeA2 3gyrL2 |
UNCERT: 1.531E-1 |
0.3041 |
 |
|
|
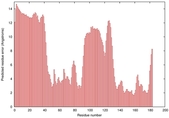
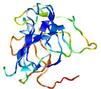
IntFOLD5 MUSTER multi4 TS1 1ov8A 6hbeA2 3gyrL2 |
UNCERT: 1.778E-1 |
0.2991 |
 |
|
|
IntFOLD5 spk2 multi4 TS1 4f2eA 1gw0A1 1j9qA1 |
UNCERT: 4.289E-1 |
0.2674 |
 |
|