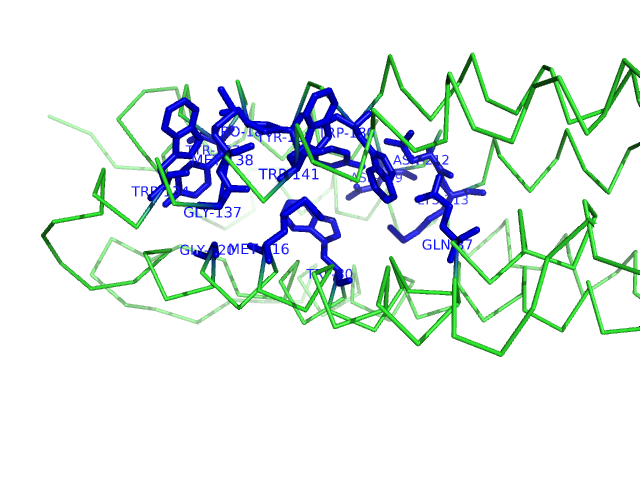
PyMOL generated image of ligand binding residues prediction for T0682
Click here to download PDB file of this model with the superposition of all identified ligands.
Predicted ligand binding residues are shown as blue sticks in the image above.
Binding site: 80, 87, 116, 120, 134, 137, 138, 141, 180, 183, 184, 187, 209, 212, 213
Most likely ligand (Type): RET
Centroid ligand (Type): RET
All ligands clustered at site (Type-Number): RET-11
JSmol view of ligand binding residues prediction for T0682
The FunFOLD2 server makes use of the FunFOLDQA Quality Assessment protocol for improved prediction selection::
Roche, D. B., Buenavista, M. T. & McGuffin, L. J. (2012) FunFOLDQA: A Quality Assessment Tool for Protein-Ligand Binding Site Residue Predictions. PLoS ONE, 7, e38219. PubMed
BDTalign = 0.7426803488573831
Identity = 0.44365266
Rescaled BLOSUM62 = 0.41487166
Equivalent Residue Ligand Distance = 0.49493766
Model Quality = 0.7159
Predicted BDT score = 0.82843095
Predicted MCC score = 0.7650769
r=TRP; n=141; | I=000; O=0.167; N=000; P=000; |
r=MET; n=116; | I=000; O=0.183; N=000; P=000; |
r=ASN; n=212; | I=000; O=0.075; N=000; P=000; |
r=GLN; n=87; | I=000; O=0.070; N=000; P=000; |
r=PRO; n=184; | I=000; O=0.154; N=000; P=000; |
r=TRP; n=134; | I=000; O=0.207; N=000; P=000; |
r=MET; n=138; | I=000; O=0.116; N=000; P=000; |
r=TYR; n=187; | I=000; O=0.086; N=000; P=000; |
r=TYR; n=183; | I=000; O=0.207; N=000; P=000; |
r=GLY; n=120; | I=000; O=0.117; N=000; P=000; |
r=ASP; n=209; | I=000; O=0.077; N=000; P=000; |
r=GLY; n=137; | I=000; O=0.121; N=000; P=000; |
r=LYS; n=213; | I=000; O=0.056; N=000; P=000; |
r=TRP; n=180; | I=000; O=0.273; N=000; P=000; |
r=TRP; n=80; | I=000; O=0.146; N=000; P=000; |
3D models for FunFOLD2 are built using the IntFOLD2-TS protocol:
Buenavista, M. T., Roche, D. B. & McGuffin, L. J. (2012) Improvement of 3D protein models using multiple templates guided by single-template model quality assessment. Bioinformatics, 28, 1851-1857. PubMed
The original paper describing FunFOLD version 1.0:
Roche, D. B., Tetchner, S. J. & McGuffin, L. J. (2011) FunFOLD: an improved automated method for the prediction of ligand binding residues using 3D models of proteins. BMC Bioinformatics, 12, 160. PubMed
